In 1977, Frederick Sanger and his team invented the chain termination DNA sequencing technology, namely Sanger sequencing technology, which opened a door for human beings to explore the genetic code of life and won the Nobel Prize again. Since its inception, Sanger sequencing technology has experienced a series of important development stages. In the early days, this technology relied on plate gel electrophoresis to separate fragments, which was inefficient, and only a few hundred bases could be determined in one experiment and the operation was cumbersome and time-consuming.
With the development of technology, capillary electrophoresis has gradually replaced plate gel electrophoresis, which has realized the automation of the sequencing process and significantly improved the sequencing speed and throughput. At the same time, the introduction of fluorescent labeling technology makes base recognition more accurate, and sequencing accuracy has greatly improved. Sanger sequencing technology has played a central role in the human genome project, providing key data for the mapping of the human genome and establishing its classic position in the field of molecular biology.
Nowadays, although high-throughput sequencing technology (such as Illumina sequencing, PacBio sequencing, etc.) dominates the whole genome sequencing, metagenomics and other fields with its ultra-high sequencing throughput, Sanger sequencing technology is still irreplaceable in many application scenarios with its unique advantages such as high accuracy, long reading time and single molecule resolution. Looking forward to the future, Sanger sequencing technology has not stagnated, but is constantly innovating and developing. Its technical performance will be further optimized, and its application fields will continue to expand, and it will continue to play an important role in life science research and clinical applications.
This article explores the technical development and innovative applications of Sanger sequencing, and looks forward to its future prospects in terms of technical optimization, cost reduction and expanded application fields.
Technological Development of Sanger Sequencing
From early plate gel electrophoresis to capillary electrophoresis automation, from manual interpretation to software-aided analysis, it keeps the advantage of high accuracy and continuously optimizes the speed and throughput. So far, it is still the gold standard for accurate sequencing of single fragments, and it continues to expand the boundaries in technical integration.
Continuous Improvement of Sequencing Performance
In recent years, researchers and technology developers have made remarkable progress in Sanger sequencing technology in terms of sequencing speed, accuracy, and throughput by continuously optimizing the experimental process and improving the performance of instruments and reagents.
In terms of sequencing speed, the new generation of automatic sequencers adopts a more efficient capillary array and faster electrophoretic separation technology, which greatly shortens the running time of single sequencing. The early Sanger sequencer took several hours to complete a sequencing reaction, but now the advanced instruments can complete the sequencing of the same length in 1-2 hours, and can process more samples at the same time. For example, some Qualcomm Sanger sequencing platforms can run 96 or 384 capillaries at the same time, which greatly increases the sequencing sample size per unit time and meets the efficient requirements of small and medium-sized sequencing projects.

(A) Changes of read length and degree of parallelism in sequencing technologies since the 1990s up to the present, (B) Number of reads and read length per sequencing technology (Stranneheim et al., 2012)
Accuracy is the core advantage of Sanger sequencing technology. At present, the single-base sequencing error rate has dropped below 0.1%, and the error rate can be further reduced to 0.01% or even lower through two-way sequencing or repeated sequencing. This progress is mainly due to the development and optimization of high-fidelity DNA polymerase and a more accurate fluorescence detection system. The new DNA polymerase has stronger proofreading activity and can effectively reduce base mismatches. However, the high-resolution optical detection system can identify the bases with different fluorescent labels more accurately and reduce the misjudgment caused by signal interference.
Besides increasing the number of capillaries, the application of microfluidic chip technology provides a new possibility for Qualcomm’s quantification of Sanger sequencing. Microfluidic chip realizes the integration and automation of sequencing reaction through miniaturized channels and reaction cells, which greatly reduces reagent consumption and reaction time, and improves the parallelism of sample processing. For example, the Sanger sequencing system, based on a microfluidic chip, can perform thousands of sequencing reactions on a chip at the same time. Although the cost of a single reaction is slightly higher than that of traditional capillary sequencing, it has significant cost-effectiveness advantages when dealing with large-scale samples.

(A) Comparison of number of DNA molecules required for generating a base call inconsensus sequencing and single-molecule sequencing, (B) The most common type ofsequencing errors per sequencing technology (Stranneheim et al., 2012)
Exploration of the Application of Emerging Technologies
With the deepening of life science research, the combination of Sanger sequencing technology and other cutting-edge technologies has spawned a series of emerging application directions and expanded its application scope.
Single-cell Sanger sequencing is one of the most concerning technologies in recent years. By sequencing the genomic DNA or RNA of a single cell, researchers can reveal the heterogeneity between cells and provide a new perspective for the research in oncology, neuroscience, and other fields. Traditional Sanger sequencing needs a lot of template DNA, but the DNA content of a single cell is extremely low (only about 6pg), which makes it difficult to meet the demand of direct sequencing.
For this reason, researchers have developed genome-wide amplification (WGA) technology, such as multiple displacement amplification (MDA), which can amplify the DNA of a single cell by millions of times while maintaining the integrity of the genome, and provide enough templates for the subsequent Sanger sequencing. For example, in tumor research, single-cell Sanger sequencing can analyze the gene mutation of different cells in tumor tissue, identify the specific mutation of tumor stem cells or drug-resistant cells, and provide a basis for personalized treatment.

The principle of single-cell sequencing (Tang et al., 2019)
The combination of nanopore sequencing and Sanger sequencing is another potential technical direction. Nanopore sequencing technology has the advantages of long reading length and real-time sequencing, but its single-base accuracy is still lower than Sanger sequencing. Combining the two technologies, the results of nanopore sequencing can be verified and corrected by using the high accuracy of Sanger sequencing, and the quality of overall sequencing data can be improved.
For example, when sequencing complex genomic regions (such as highly repetitive sequences and GC-rich regions), firstly, nano-pore sequencing is used to obtain long reading sequences, covering the whole region, and then Sanger sequencing is used to verify key regions or suspicious mutation sites, which not only ensures the integrity of sequencing, but also ensures the accuracy of the results.
In addition, the application of Sanger sequencing technology in the field of in situ sequencing is also being explored. In-situ sequencing can detect the position and expression level of specific nucleic acid sequences in tissue slices or cell samples in situ, and retain the spatial structure information of the samples. By combining the principle of Sanger sequencing with in situ hybridization, the spatial distribution of specific genes in tissues can be analyzed with high resolution by sequencing reaction in cells and detecting fluorescence signals, which provides a new tool for developmental biology, pathology, and other research.

The clinical applications of single-cell sequencing in different fields, like cancer, immunesystem, reproductive system, microorganisms, etc (Tang et al., 2019)
Sanger sequencing can be provided by CD Genomics. As the "gold standard" of gene detection, is famous for its ultra-high accuracy at single base level, with a stable reading length of 500-800bp. It is the first choice for mutation confirmation, gene editing verification, plasmid sequencing, and provides irreplaceable accurate data support for scientific research.
Sanger Sequencing Innovative Application
Sanger sequencing, with its unique advantage of high accuracy, is breaking through the traditional application boundary and rejuvenating. From accurate verification of gene editing to sequence quality control of synthetic biology, from mutation detection to single cell heterogeneity analysis, its innovative application in frontier fields has opened up a more accurate path for life science research and clinical practice.
Gene Editing Verification
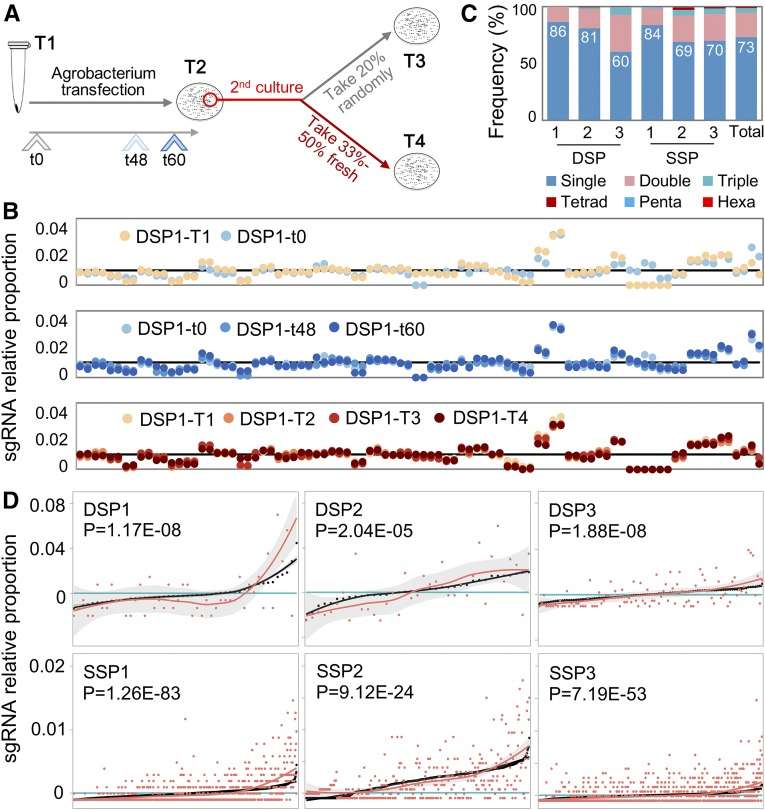
The rapid development of gene editing technology (such as CRISPR-Cas9) has brought revolutionary breakthroughs in gene function research and gene therapy, and Sanger sequencing technology is the gold standard to verify the efficiency and accuracy of gene editing. After using CRISPR-Cas9 technology to knock out a gene, researchers amplify the target gene region by PCR and then perform Sanger sequencing. By comparing with the wild-type sequence, we can accurately detect whether the gene editing is successful, calculate the editing efficiency, and determine the mutation type (such as insertion, deletion, or point mutation).

Pipeline of High-Throughput Genome-Editing Design (Liu et al., 2020)
Synthetic Biology
In the field of synthetic biology, Sanger sequencing technology is widely used for verification and quality control of synthetic genes and genomes. Synthetic gene fragments may introduce errors in the process of splicing and assembly, and each synthetic fragment can be verified one by one by Sanger sequencing to ensure that its sequence is consistent with the design.
Combination with Other Cutting-Edge Technologies
The combination of Sanger sequencing technology and other cutting-edge technologies brings more opportunities and possibilities for biological research and applications.
The complementary application of gene chip technology and Sanger sequencing technology is of great value in clinical diagnosis. Gene chip can be used to screen large-scale gene mutations and quickly find possible mutation sites, while Sanger sequencing can accurately verify the positive results detected by the chip, eliminate false positives, and improve the accuracy of diagnosis.
The combination of Sanger sequencing and mass spectrometry can be used to verify the amino acid sequence in protein genomics research. Protein was hydrolyzed to obtain a peptide, and the molecular weight and sequence information of the peptide were determined by mass spectrometry. Then the gene encoding protein was sequenced by Sanger sequencing, which could realize the mutual verification between the gene sequence and the protein sequence and improve the accuracy of protein identification.
High Coverage and Uniformity from Plasmid Pool to To Individuals (Liu et al., 2020)
Future Prospects of Sanger Sequencing
Despite the rapid development of high-throughput sequencing technology, Sanger sequencing is still irreplaceable in the field of accurate detection because of its high accuracy of single base and long reading time. In the future, it will evolve to higher speed, lower cost, higher automation, integrate with microfluidic and nanotechnology, expand in clinical verification, single cell analysis, and other scenarios, and continue to provide core support for life science research and precision medicine.
In terms of technical performance, sequencing speed will usher in a revolutionary breakthrough. Researchers are exploring the combination of microfluidic chip technology and traditional electrophoretic separation technology, which can greatly shorten the separation time by realizing the rapid migration of DNA fragments in nano-scale channels.
At the same time, the new generation of ultra-sensitive fluorescence signal detection system will use single photon detector and artificial intelligence algorithm to process weak signals in real time, so that the running time of single sequencing is expected to be shortened from the current several hours to less than 30 minutes, and the base recognition accuracy is ensured to be ≥99.9%, while the background noise interference is significantly reduced.
In addition, the sequencing reaction will be more automated, and the whole process from sample processing, reaction system construction to result analysis will be integrated and intelligent, reducing human operation errors and improving experimental efficiency.

Applied Targeted Mutagenesis for the Validation of Gene Function (Liu et al., 2020)
Cost reduction is another important trend of Sanger sequencing technology in the future. At the level of reagent research and development, researchers are transforming DNA polymerase through directed evolution technology, and optimizing the expression system of high-fidelity enzyme by using Qualcomm screening platform, so that the production cost is reduced by more than 40%.
In the field of instrument manufacturing, 3D printing technology is applied to the production of key components, and the processing cost of precision components is reduced through customized design. Modular instrument architecture allows users to flexibly combine functional modules according to their needs, reducing unnecessary equipment investment.
In addition, the popularity of cloud computing services has greatly reduced the cost of data analysis. Users can obtain professional-level sequence analysis services through subscription mode without purchasing high-performance local servers. It is expected that the single reaction cost will drop to 1/3 of the current level in the next five years.
In terms of application field expansion, Sanger sequencing technology will play a greater role in food safety, environmental monitoring, and other fields. In the field of food safety, Sanger sequencing can be used to detect foreign gene sequences in genetically modified foods, identify species components in foods, and prevent counterfeit and shoddy products. In environmental monitoring, the structure and changes of microbial communities can be monitored by Sanger sequencing of microbial DNA in environmental samples, and the environmental quality and pollution degree can be evaluated.
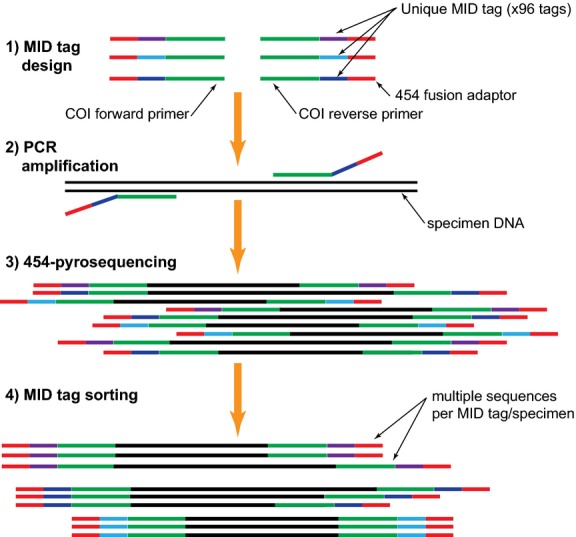
Schematic diagram of parallel barcode recovery using multiple identifier (MlD) tagging and next-generation sequencing (NGS) protocol (Shokralla et al., 2014)
Conclusion
After decades of development, Sanger sequencing technology still plays an important role in life science research and clinical applications since its invention. Its high accuracy, long reading time, and reliability make it irreplaceable in the fields of gene editing and verification, synthetic biology, personalized medicine, and so on. Despite the competition from high-throughput sequencing technology, Sanger sequencing technology has been continuously revitalized through continuous technological innovation and application expansion.
In the future, with the further optimization of technology and cost reduction, Sanger sequencing technology will be more widely used and play an important role in solving global challenges. We encourage researchers, clinicians, and practitioners in related industries to continue to pay attention to the latest development of Sanger sequencing technology, actively explore its innovative applications in their respective fields, and make full use of this classic technology to serve scientific research and social development. It is believed that Sanger sequencing technology will continue to make important contributions to the progress of life science and human welfare through continuous innovation and practice.
Learn More
References
- Stranneheim H, Lundeberg J. "Stepping stones in DNA sequencing." Biotechnol J. 2012 7(9): 1063-1073 https://doi.org/10.1002/biot.201200153
- Tang X, Huang Y, Lei J, Luo H, Zhu X. "The single-cell sequencing: new developments and medical applications." Cell Biosci. 2019 9: 53 https://doi.org/10.1186/s13578-019-0314-y
- Liu HJ, Jian L, Xu J, et al. "High-Throughput CRISPR/Cas9 Mutagenesis Streamlines Trait Gene Identification in Maize." Plant Cell. 2020 32(5): 1397-1413 https://doi.org/10.1105/tpc.19.00934
- Shokralla S, Gibson JF., et al. "Next-generation DNA barcoding: using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens." Mol Ecol Resour. 2014 14(5): 892-901 https://doi.org/10.1111/1755-0998.12236


 Sample Submission Guidelines
Sample Submission Guidelines