PCR-LDR SNP Genotyping Service
With the advancement of genomics and molecular biology techniques, high-throughput molecular marker technologies such as SNP chips and Genotyping-By-Sequencing (GBS) based on second-generation sequencing have been developed. These technologies hold considerable promise across a spectrum of research areas, including the identification of SNP loci among diverse genetic materials, the selection of genetic backgrounds in crop breeding, and genome-wide association studies. However, their application in molecular marker-assisted breeding for crops is hindered by the relatively high costs and complexity of the experimental procedures involved.
In the practical implementation of molecular marker-assisted breeding for crops, numerous functional polymorphic loci associated with key agronomic traits are often employed. To this end, molecular markers founded on PCR technology are devised for these loci, thereby facilitating the assisted selection of genetic lineages.
What is Ligase Detection Reaction (LDR) Technology
Ligase Detection Reaction (LDR) technology is a highly precise molecular technique employed for the identification of single nucleotide polymorphisms (SNPs) in DNA. This technique hinges on the use of Taq DNA ligase, an enzyme that facilitates the ligation of oligonucleotide probes. Ligation occurs exclusively when these probes are perfectly complementary to the target DNA sequence, ensuring continuity without gaps. In the presence of mismatches, such as base substitutions or insertions/deletions, the ligation does not proceed.
The LDR process involves two principal steps: the hybridization of probes to the target DNA and the subsequent ligation of these probes upon achieving a perfect match. Following ligation, fluorescence scanning and gel electrophoresis are employed to analyze fragment sizes and detect variations in the DNA sequence. This technique affords high resolution and specificity in SNP detection by scrutinizing base differences at the probe termini.
Introduction of PCR-LDR SNP Genotyping Service
CD Genomics offers an advanced PCR-LDR SNP genotyping service designed to deliver high-precision genetic analysis. This service combines polymerase chain reaction (PCR) with LDR technology to accurately identify SNPs across multiple loci concurrently. The integration of these methods ensures robust performance in SNP genotyping, particularly in the context of diverse and complex genetic samples.
Our PCR-LDR service harnesses the high specificity of LDR for detecting minute genetic variations, alongside the amplification capability of PCR. This approach is well-suited for a broad spectrum of applications, spanning from basic research to applied genetic studies, and ensures reliable and accurate results.
Methods of SNP Genotyping
Sequenom MassArray
The Sequenom MassArray method employs mass spectrometry to detect SNPs by measuring the mass of PCR products. While it is highly accurate, the method can be costly and is typically reserved for high-throughput analyses.
Multiplex SNaPshot SNP Genotyping
This technique uses fluorescently labeled dideoxynucleotides to determine SNP genotypes. It facilitates multiplex analysis of multiple SNPs in a single reaction. However, the interpretation of resulting data can be complex, requiring advanced analytical skills.
LDR (PCR-LDR)
LDR technology, as implemented in our PCR-LDR service, involves the utilization of a high-temperature ligase enzyme to recognize SNPs by ligating perfectly matching probes. This method ensures exceptional accuracy and can be adapted to various SNP loci, making it suitable for both low- and medium-throughput analyses.
Direct Sequencing
Direct sequencing provides detailed information by sequencing the DNA directly. Although this method is highly accurate, it tends to be more resource-intensive compared to other genotyping techniques, often limiting its application to specific contexts where such detail is essential.
Advantages of PCR-LDR SNP Genotyping Service
- Universality: Applicable for the genotyping of any polymorphic locus and various SNP loci, without the need for the introduction of restriction enzyme sites.
- Accurate Typing with High Detection Rates: Accuracy can exceed 99.2%.
- Rapid Detection: The most significant advantage of LDR technology is its short experimental time. Compared to RFLP, SSCP, and DHPLC, LDR technology allows for easier control of detection conditions, resulting in an overall experimental duration of only a few weeks.
- Flexible Experimentation, High Detection Rates, and Short Experimental Cycles.
- Double assurance based on hybridization and connection reactions, ensuring accurate results.
- Stringent quality control during the experimental process and a 10% replicate control, ensuring result accuracy.
- Suitable for medium to low SNP typing scales, recommended for use with up to 30 SNP loci and sample sizes of fewer than a thousand.
Applications of PCR-LDR SNP Genotyping
This detection technology is well-suited for various medium- to low-throughput SNP testing scales, including quantitative trait loci (QTL) localization studies, candidate gene or locus association analysis, molecular breeding, and other related applications.
- Genetic Research: PCR-LDR SNP genotyping is essential in genetic research for identifying genetic variations associated with traits or diseases. It supports studies in population genetics, evolutionary biology, and functional genomics.
- Molecular Breeding: In agriculture, our PCR-LDR service facilitates molecular breeding by identifying SNPs associated with desirable traits. This application aids in developing new plant varieties with enhanced characteristics.
- Clinical Genetics: The service is valuable for clinical research and diagnostics, helping to identify genetic markers related to various diseases and conditions. It supports personalized medicine by providing insights into genetic predispositions and therapeutic targets.
- Forensic Analysis: PCR-LDR technology is useful in forensic science for analyzing degraded DNA samples, such as those found at crime scenes. Its sensitivity and accuracy make it an effective tool for forensic investigations.
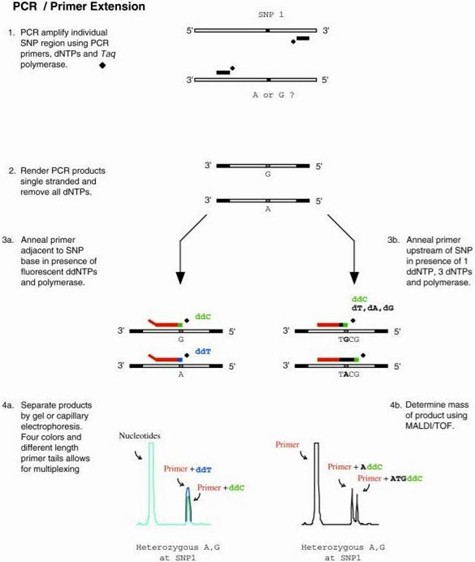
PCR-LDR SNP Genotyping Technology Workflow
- Multiplex PCR Amplification: Obtaining the fragment where the target locus is located.
- Multiplex LDR: Producing detection products of varying lengths.
- Sequencing: Different peaks are obtained based on sequencing electrophoresis.
- Interpretation of SNP results and data analysis.
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- Experimental results
- Data analysis report
- Details in PCR-LDR SNP Genotyping for your writing (customization)
CD Genomics offers a complete PCR-LDR SNP Genotyping Service, managing everything from sample preparation and PCR amplification to accurate SNP detection and analysis. We provide custom probe design and tailored solutions to fit your research needs. For more details or to discuss your project, please contact our team.
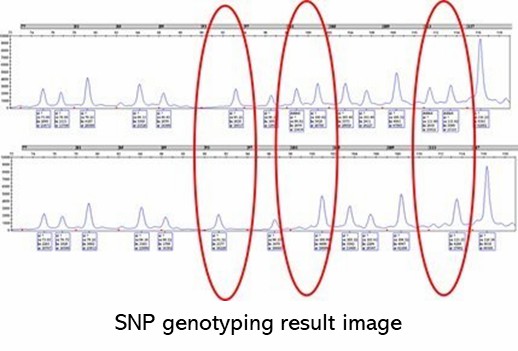
Demo Results
Partial results are shown below:
PCR-LDR SNP Genotyping Service FAQs
1. What types of samples can be analyzed using the PCR-LDR SNP genotyping service?
The PCR-LDR SNP genotyping service is adept at analyzing a diverse range of sample types, including blood, tissue, and saliva. Its efficacy extends to degraded DNA samples, showcasing the robustness of the PCR-LDR technology across various sample conditions.
2. How many SNPs can be analyzed in a single experiment?
Our service is equipped to analyze up to 30 SNPs in a single experiment. This capacity renders it highly suitable for medium-scale projects and extensive genetic studies that demand precise SNP analysis.
3. What is the turnaround time for results?
Generally, results are furnished within a few weeks. This expedited turnaround time is attributed to the efficient workflow and rapid processing capabilities intrinsic to our PCR-LDR service, ensuring timely delivery of genetic data.
4. How is the accuracy of the results ensured?
To guarantee accuracy, stringent quality control measures are implemented, including the re-analysis of 10% of samples. Furthermore, the high precision of the LDR technology significantly bolsters the reliability of the results.
PCR-LDR SNP Genotyping Service Case Studies
The association between pre-miR-27a rs895819 polymorphism and myocardial infarction risk in a Chinese Han population
Journal: Lipids in health and disease
Impact factor: 4.5
Published: 06 January 2018
Background
Myocardial infarction (MI) is a significant health challenge in China, involving both environmental and genetic factors. This study utilized PCR-LDR to perform SNP genotyping on 287 MI patients and 646 controls. It identified that the pre-miR-27a rs895819 SNP is associated with MI susceptibility in the Chinese Han population, likely affecting HDL-C levels.
Materials & Methods
Sample Preparation:
- Human
- Peripheral blood sample
- DNA extraction
Method:
- SNP genotyping
- PCR-LDR method
Data Analysis:
- Hardy-Weinberg equilibrium
- Logistic regression analysis
- One-way analysis of variance (ANOVA)
Results
Genotype Distribution: Genotype distributions followed Hardy-Weinberg equilibrium in controls, indicating no population stratification.
Table 1 Multivariate associations of the pre-miR-27a rs895819 with the risk of MI
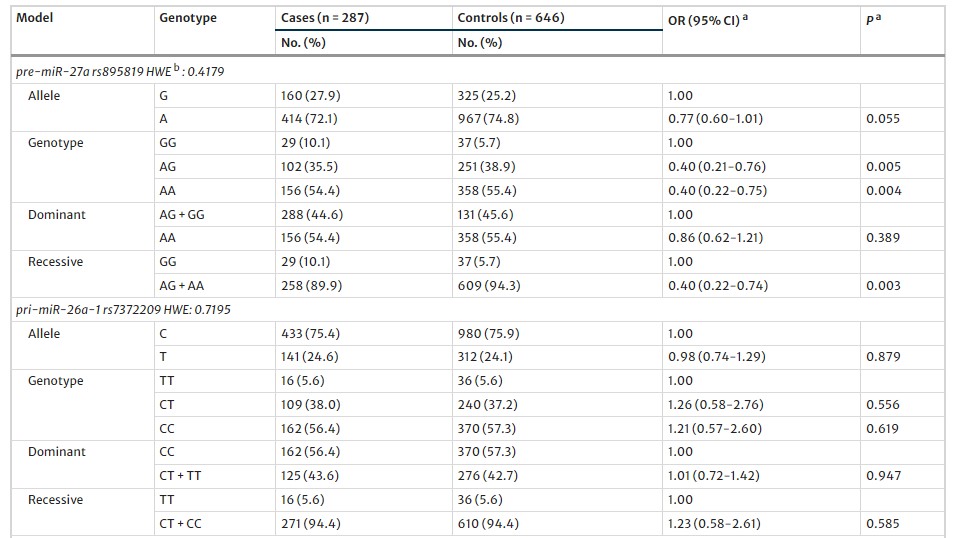
Association with MI Risk: The AG and AA genotypes of pre-miR-27a rs895819 were linked to reduced MI risk compared to the GG genotype. Other miRNA polymorphisms showed no significant association.
Table 2 Multivariate associations of the pre-miR-27a rs895819 with the risk of MI by further stratification for age, gender, smoking and drinking
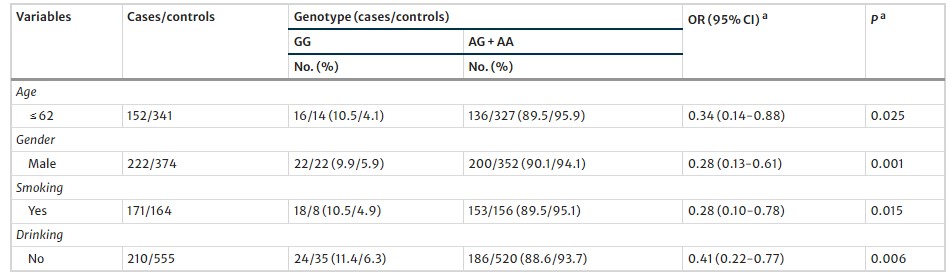
Stratified Analysis: The reduced MI risk associated with AG+AA genotypes was more evident in younger individuals, males, and smokers.
Table 3 ANOVA analysis of the association between pre-miR-27a rs895819 and the TG, TC, LDL-C and HDL-C levels
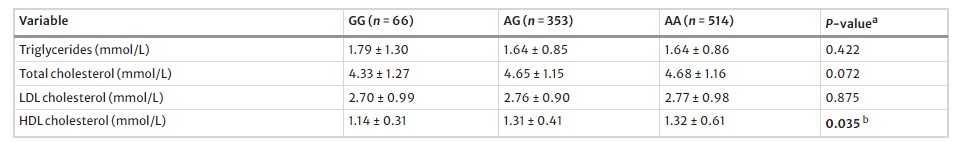
Lipid Profile: AG and AA genotypes of pre-miR-27a rs895819 were associated with higher HDL-C levels compared to the GG genotype.
Conclusion
In conclusion, the AG and AA genotypes of pre-miR-27a rs895819 are linked to a reduced risk of MI in the Chinese Han population, especially among younger individuals, males, and smokers. This association may be related to higher HDL-C levels. Further research is needed to validate these findings and understand the underlying mechanisms.
Reference:
- Cai MY, Cheng J, Zhou MY, Liang LL, Lian SM, Xie XS, Xu S, Liu X, Xiong XD. The association between pre-miR-27a rs895819 polymorphism and myocardial infarction risk in a Chinese Han population. Lipids in health and disease. 2018, 17:1-8.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Use of biostimulants for water stress mitigation in two durum wheat (Triticum durum Desf.) genotypes with different drought tolerance
Journal: Plant Stress
Year: 2024
The Restriction-Modification Systems of Clostridium carboxidivorans P7
Journal: Microorganisms
Year: 2023
In the land of the blind: Exceptional subterranean speciation of cryptic troglobitic spiders of the genus Tegenaria (Araneae: Agelenidae) in Israel
Journal: Molecular Phylogenetics and Evolution
Year: 2023
Genetic Modifiers of Oral Nicotine Consumption in Chrna5 Null Mutant Mice
Journal: Front. Psychiatry
Year: 2021
A high-density genetic linkage map and QTL identification for growth traits in dusky kob (Argyrosomus japonicus)
Journal: Aquaculture
Year: 2024
Genomic and chemical evidence for local adaptation in resistance to different herbivores in Datura stramonium
Journal: Evolution
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
