Microsatellite Genotyping Service
CD Genomics has extensive experience in providing support for the selection and design of microsatellite markers for a wide range of plant and animal species. Besides microsatellite genotyping, we also provide genotyping by sequencing (GBS) method, which saves time and resources.
The Introduction of Microsatellite Genotyping
Microsatellites, also known as simple sequence repeats (SSRs) or short tandem repeats (STRs), have been popular markers due to their high polymorphism. Genotyping is an accurate, cost-effective, and fast approach to distinguish microsatellite alleles, boosted by successive technical advances, including multiplexing PCR and next-generation sequencing technologies. The complete workflow for microsatellite genotyping includes acquisition of sequencing data, SSR selection, primer design, primer validation, multiplexing PCR, and capillary gel electrophoresis coupled with fluorescence-based detection, as well as data analysis.
The PCR reaction is performed with fluorescent dye-labeled primers, then the PCR fragments can be analyzed on a capillary DNA sequencing machine, and the data is analyzed using GeneMapperTM software. By taking advantage of multiplexing by fragment size and dye color, and the ability of automated fluorescent genetic analyzer machines to autoload 16 samples at a time from 96-well or even 384-well microtiter plates, high-throughput marker analysis can be designed. This service has been applied to a wide variety of species and can accommodate any species for which microsatellite markers are available.
If you would like to learn more about Microsatellite Genotyping, you can read our article "Introduction to Microsatellite and Microsatellite Genotyping".
Advantages of Microsatellite Genotyping
Advantages of microsatellites as genetic markers:
- Universal and polymorphic
- Locus-specific (in contrast to multi-locus markers such as minisatellites)
- Codominant (heterozygotes can be distinguished from homozygotes)
- PCR-based (only requires tiny amounts of tissue and works on degraded DNA)
- Multiple applications (from individual identification to fine-scale phylogenies)
Application of Microsatellite Genotyping
The vast amount of data emerging for thousands of microsatellite markers across organisms makes microsatellite genotyping a widely accepted tool for:
- Linkage analysis of co-segregation
- Diagnosis and identification of human diseases
- Forensic identification and relatedness testing
- Cell line identification
- Population studies
Microsatellite Genotyping Workflow
CD Genomics adopts advanced platforms (ABI 3730xl DNA Analyzer) and strategies (multiplexing PCR) to provide a fast and accurate microsatellite genotyping service and bioinformatics analysis. Our highly experienced expert team executes quality management, following every procedure to ensure confident and unbiased results. The rough workflow is outlined below.
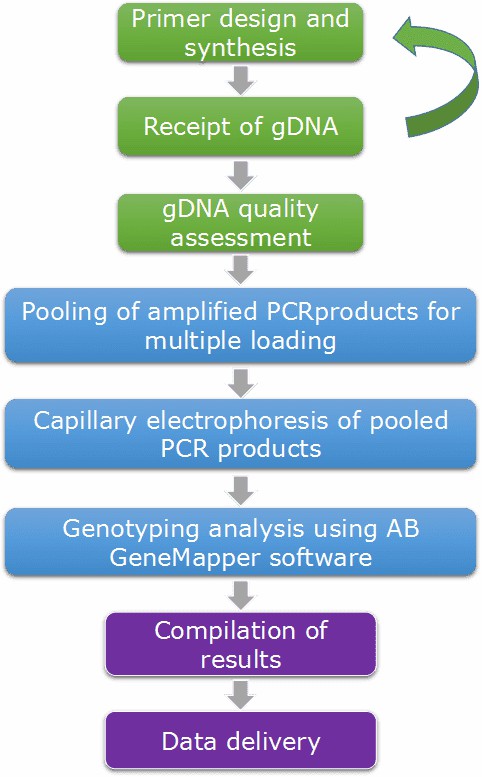
Service Specification

|
Sample Requirements
|
 |
Multiplexing PCR and Genotyping
|

|
Data Analysis
|
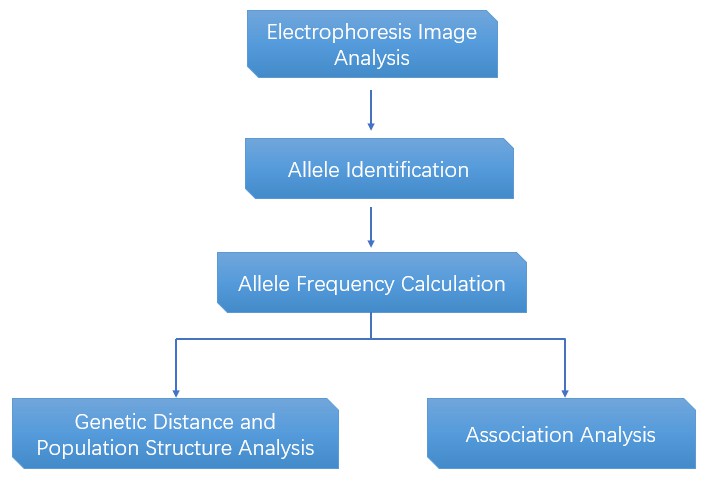
Analysis Pipeline
Deliverables
- Complete experimental procedure,
- PCR electrophoresis images
- Large gel detection images
- Sequencing chromatograms
- Data analysis results
CD Genomics provides full microsatellite genotyping service package, including designing and ordering fluorescently labeled primer pairs, primer validation, microsatellite genotyping, as well as data analysis. We can tailor this pipeline to your research interest. If you have additional requirements or questions, please feel free to contact us.
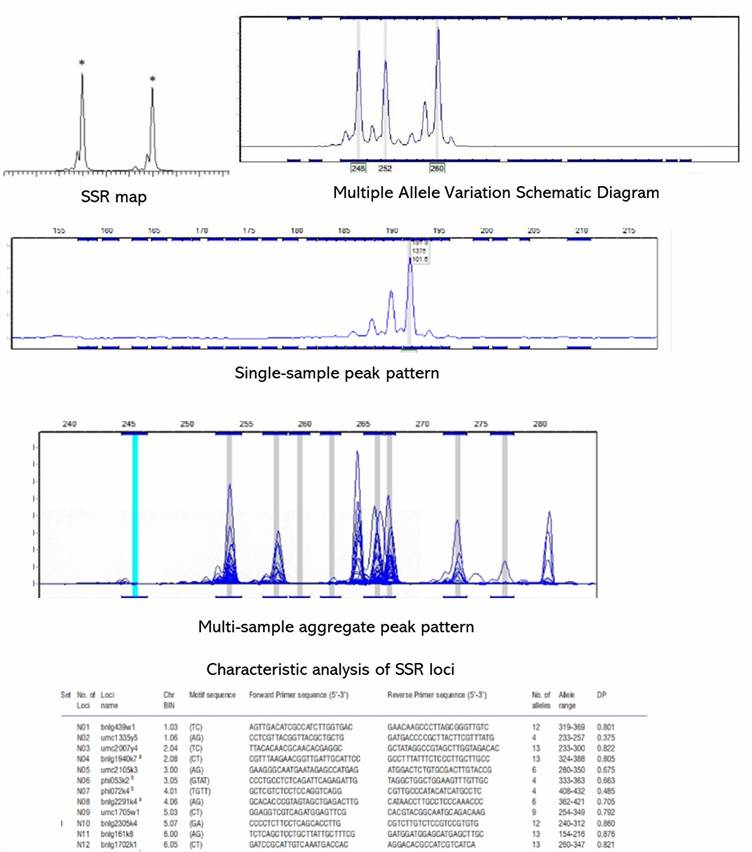
Demo Results
Microsatellite Genotyping Service FAQs
1. What is Microsatellite Genotyping?
Microsatellite Genotyping is a powerful technique employed for the detection and evaluation of variations within microsatellites, also known as Simple Sequence Repeats (SSRs), in DNA. Microsatellites are short repeating sequences composed of 1-6 base pairs. These sequences are considerably variable across the genome, hence, they serve as excellent genetic markers.
2. What is the Principle Underlying Microsatellite Genotyping?
Microsatellite Genotyping is grounded in the principle of Polymerase Chain Reaction (PCR) for the amplification of specific microsatellite sites. This technique entails the design of specific primers to amplify targeted microsatellite regions. Subsequently, the amplified products are differentiated and typed based on size variations via techniques such as gel electrophoresis or capillary electrophoresis.
3.What are the advantages of Microsatellite Genotyping Technology?
The use of microsatellite genotyping technology offers several distinct advantages in studies of genetic variation:
- Prominent Polymorphism: Microsatellites are characterized by their high degree of variability, thereby supplying an abundance of genetic information.
- Precise Resolution: This investigative technology enables highly precise differentiation, teasing out even minute genetic variations among individual organisms.
- Broad Distribution: Microsatellite markers pervade the genome extensively, which renders this technique suitable for different biological species.
4. How Does Microsatellite Genotyping Differ from Other Genotyping Technologies?
Microsatellite genotyping is distinct from Single Nucleotide Polymorphism (SNP) genotyping in multiple critical aspects:
- Enhanced Polymorphism: When compared to SNP genotyping, microsatellite genotyping features a superior level of polymorphism, making it exceptionally equipped for the exploration of highly variable genetic regions.
- Differing Application Purposes: While SNP genotyping conventionally finds utility in analysing point mutations, microsatellite genotyping excels in studies that necessitate a more detailed understanding of genetic variability.
5. How can microsatellite genotyping data be processed and analyzed?
The procedures for processing and analyzing microsatellite genotyping data encompass:
- Fragment size determination: Measurement of fragment sizes from electropherograms utilizes specialized software.
- Genotyping: Based on fragment size, identification of alleles at microsatellite loci occurs.
- Statistical analysis: Genotypic data is analyzed with statistical software to expose genetic variability and population structure.
Microsatellite Genotyping Service Case Studies
High-depth, high-accuracy microsatellite genotyping enables precision lung cancer risk classification
Journal: Oncogene
Impact factor: 7.519
Published: 31 July 2017
Background
Microsatellites are a series of tandem DNA sequences comprising short repetitive units (1-6 bp). Recent research has highlighted the pivotal role these microsatellites play in the intricate process of genetic development across a myriad of cancer types. The primary objective of this study is to identify certain microsatellite markers applicable to risk stratification for lung cancer. This would be accomplished by comparing germline exome sequences between lung cancer patients and a normative control group.
Methods
- 266 LUAD and 222 LUSC germline exome samples
- 1kGP non-tumor control samples
- Genomic DNA extraction
- Target enrichment
- Target sequencing
- Genotyping
- ROC Curve Analysis
- Mechanistic analysis
- Pathway Analysis
Results
The methodology adopted is primarily divided into two stages, (i) identification of Microsatellite (MST) sites, and (ii) validation of their status as significant markers. Utilizing data from The Cancer Genome Atlas (TCGA) consisting of 488 non-small cell lung cancer germline tumor samples, and from the 1000 Genomes Project having 399 non-cancer control samples, the first phase concentrated on distinguishing genetic variance in MST sites. This analysis led to the unveiling of 119 significant MST sites, suggesting a crucial genotype difference between cancerous and control samples. Employing similar analytical methods identified an additional 144 MST markers within other cancer types. Transitioning to the second stage involved designing a custom target enrichment kit housing 263 (119+144) MST markers, supplemented by 84 control MST markers for targeted capture sequencing. Conducting deep sequencing of 30 lung cancer samples and 89 non-cancer control samples, with an average sequencing depth of 579x ± 315, illuminated pronounced differences within 21 (13+8) MST markers among the 263 under investigation.
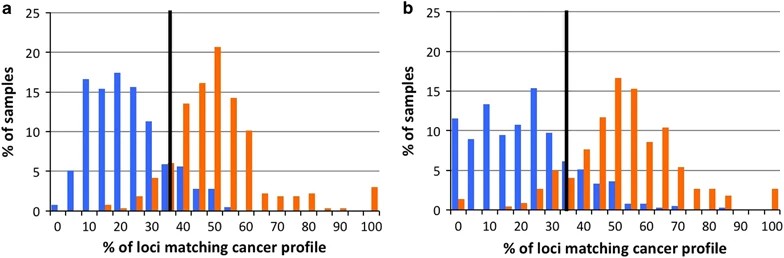 Fig 1. The computationally harvested LUAD and LUSC MST loci differentiate their corresponding cancer type from 1000 genomes non-cancer control samples with high sensitivity (LUAD: 0.87, LUSC: 0.88).
Fig 1. The computationally harvested LUAD and LUSC MST loci differentiate their corresponding cancer type from 1000 genomes non-cancer control samples with high sensitivity (LUAD: 0.87, LUSC: 0.88).
Recent studies highlight that differing cancer types can indeed share common oncogenic feature spectra. Besides discovering 13 microsatellite (MST) sites conducive to lung cancer risk stratification amid lung cancer and non-cancer data, this study also screened for 144 MST sites from other cancer types inclusive of breast, ovarian cancer, melanoma, and neuroblastomas. Following rigorous verification, it was discerned that 8 of these markers could be harnessed effectively within the purview of lung cancer risk assessment.
Table 1 MST loci that can precisely differentiate between the lung cancer samples and non-tumor samples
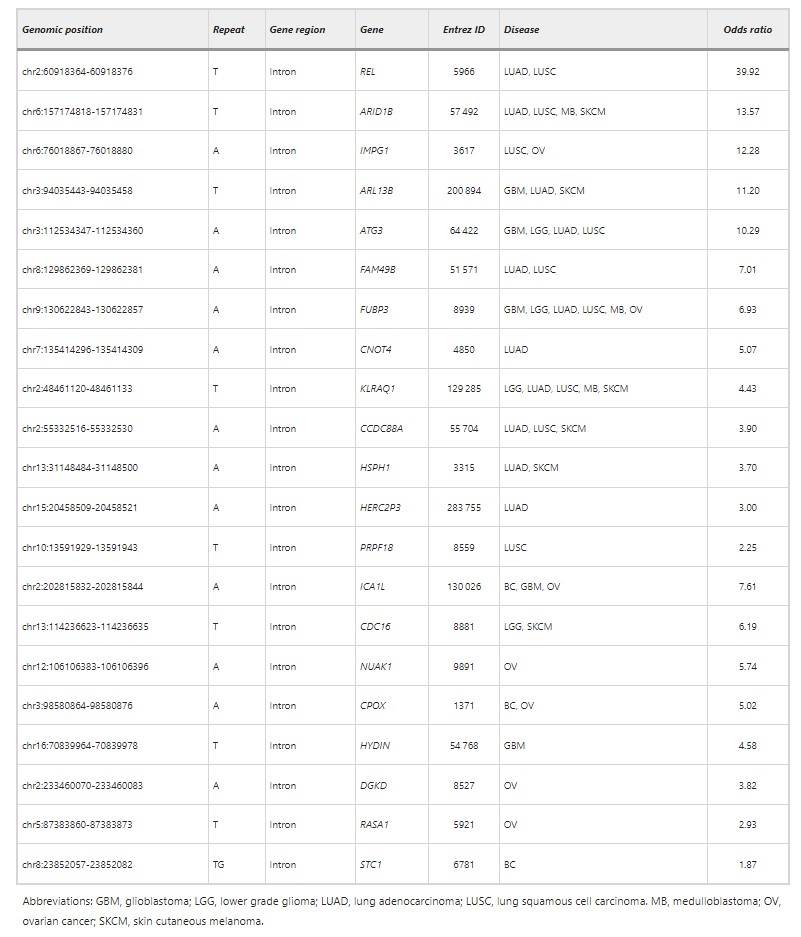
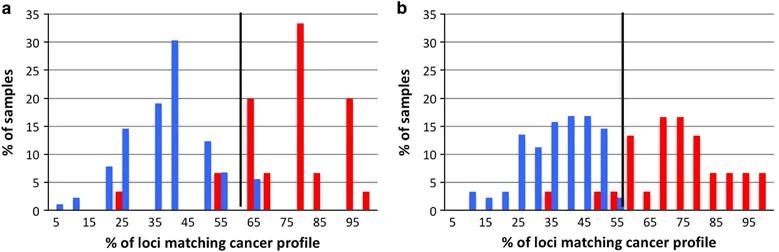 Fig 2. The 13 lung cancer specific MST loci and 8 MST loci specific for other diseases can differentiate between the lung cancer and non-cancer control sample groups.
Fig 2. The 13 lung cancer specific MST loci and 8 MST loci specific for other diseases can differentiate between the lung cancer and non-cancer control sample groups.
Further scrutiny of these 13 MST sites disclosed their location within the intron region of genes. Preceding investigations posited that intron region MST could influence the process of splice variant and transcription of genes. Cluster analysis of these 13 genes within the DAVID database revealed a snapshot of significant enrichment both in splice variant and transcript product. An association analysis of these 13 genes linked to lung cancer indicated that only REL and ARID1B were identified in previous research as being related to cancer. Implications by TCGA suggest these two genes could precipitate the occurrence of lung cancer by mechanisms of DNA damage and chromatin remodelling.
Conclusion
- A comprehensive analysis of cancer-related data from TCGA and the 1000 Genomes Project captured 263 (119+144) MST sites expressing significant genotype variations amidst cancer and control samples.
- By employing targeted capture sequencing, an effective sample validation for these MST sites was conducted that unveiled 21 (13+8) MST markers capable of facilitating superior lung cancer risk stratification.
- Subjecting the screened 13 MST sites to cluster and enrichment analysis, it was inferred that they might be influencing DNA damage and chromatin remodeling mechanisms consequently causing lung cancer. This effect is predominantly through REL and ARID1B.
- The uncovered MST sites hold potentially significant clinical value. Their discovery is paramount in the identification of new therapeutic targets, prediction of lung cancer and substantial cancer risk assessment processes.
Reference:
- Velmurugan KR, Varghese RT, Fonville NC, et al. High-depth, high-accuracy microsatellite genotyping enables precision lung cancer risk classification. Oncogene. 2017, 36(46):6383-90.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Fungi: friends or foes—an outreach science initiative for the collection of airborne fungal spores by high school students
Journal: Journal of Microbiology and Biology Education
Year: 2024
Small but significant genetic differentiation among populations of Phyllachora maydis in the midwestern United States revealed by microsatellite (SSR) markers
Journal: bioRxiv
Year: 2023
The genetic legacy of fragmentation and overexploitation in the threatened medicinal African pepper-bark tree, Warburgia salutaris
Journal: Scientific Reports
Year: 2020
Evaluation of Plasma Biomarkers for A/T/N Classification of Alzheimer Disease Among Adults of Caribbean Hispanic Ethnicity
Journal: JAMA Network Open
Year: 2023
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
