Targeted RNA Sequencing
Targeted RNA sequencing represents a state-of-the-art service provided by prominent genomics providers like CD Genomics. Researchers utilizing targeted RNA-seq can achieve unparalleled insights into fusion genes, discerning both established and previously unrecognized fusion events at a high level of detail. Such capabilities are instrumental in oncological investigations, facilitating customized therapeutic approaches and deepening our comprehension of cancer pathogenesis.
The Introduction of Targeted RNA Sequencing
Targeted RNA-sequencing selects and sequences subset transcripts of interests, increasing the coverage of a focused set of RNA sequences. It is an accurate method that generates genomic and gene-expression information of specific genome regions. The specific RNA sequences analysis enables RNA-Seq with high sensitivity and cost-effective access to NGS. By either targeted capture enrichment or amplicon-based approaches, Targeted RNA-Seq measures dozens to thousands of targets simultaneously. Enrichment assays also is a tool to identify both known and novel gene fusion partners in many sample types.
Providing qualitative and quantitative information for differential expression analysis, allele-specific expression measurement, and gene fusion verification, targeted RNA-sequencing can help to understand tumor classification and progression, genetic diseases and RNA drug response. Cancer transcriptome sequencing provides valuable information about gene expression changes in tumors. Profiling RNA-based drug response biomarkers aids too uncover multiple drug-susceptible tumorigenic pathways and improve the efficiency and success rate of the drug development process.
Advantages of Targeted RNA Sequencing
- Provides a comprehensive analysis of the specific transcripts
- Choose validated pathway, cell, or disease-specific panels
- Compatible with low-quality, formalin-fixed, paraffin-embedded (FFPE)
- Fast turnaround time and Highest data quality
- Strand information on RNA transcripts
- Effective transcriptome and pathways analysis
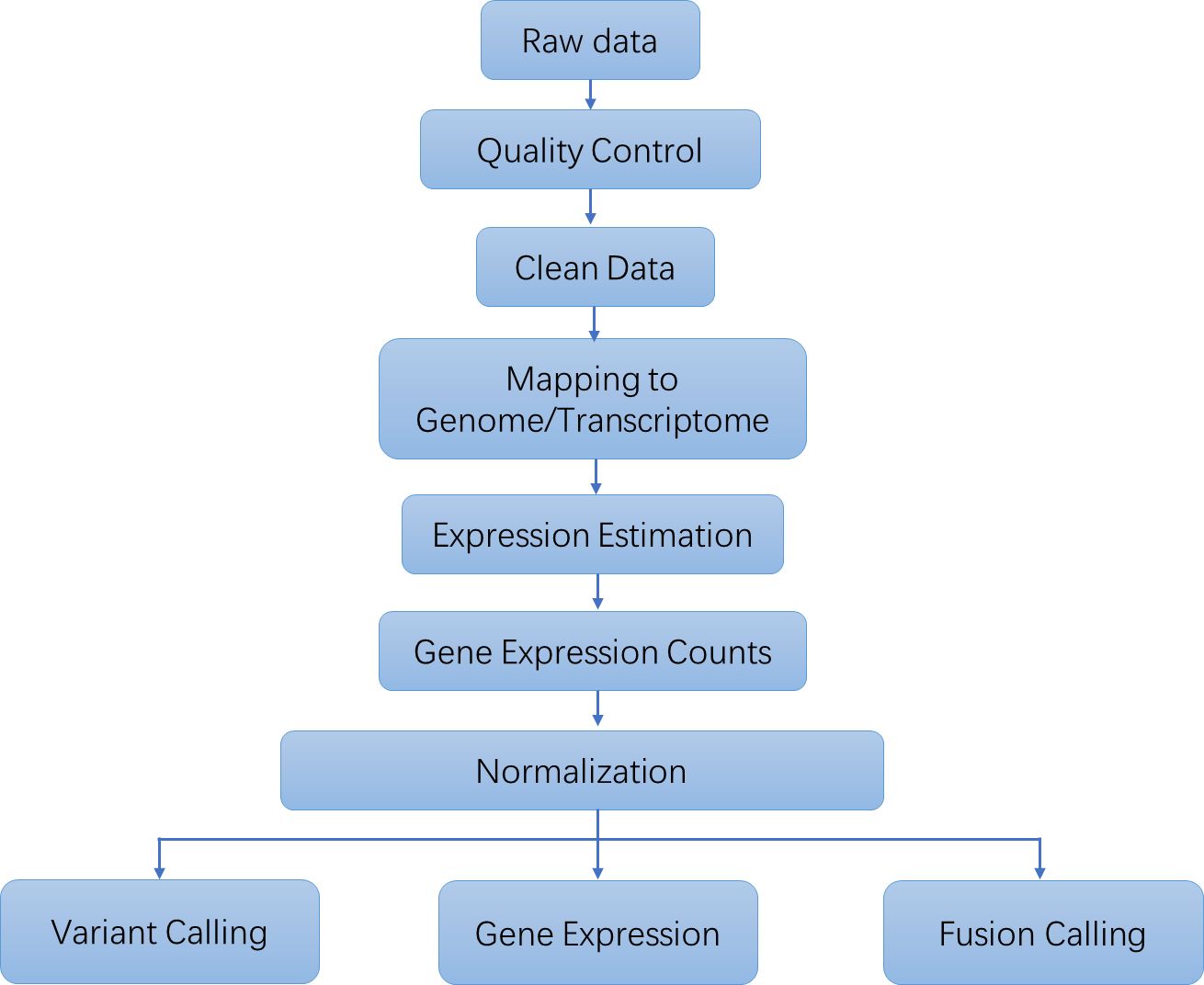
Targeted RNA Sequencing Workflow
We offer integrated targeted RNA-Seq workflows that simplify the entire process, from library preparation to data analysis and biological interpretation.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Targeted RNA Sequencing for your writing (customization)
Supported by our team of experienced scientists and advanced technology, CD Genomics offers Targeted RNA Sequencing services. We employ rigorous quality control measures and advanced bioinformatics analyses to ensure accurate and comprehensive results. If you have any specific requirements or questions, please do not hesitate to reach out to us for further assistance.
Demo Results
Partial results are shown below:

Sequencing quality distribution

A/T/G/C Distribution

IGV Browser Interface

Correlation Analysis Between Samples

PCA Score Plot

Venn Diagram

Volcano Plot

Statistics Results of GO Annotation

KEGG Classification
Targeted RNA Seq FAQs
1. How does targeted RNA sequencing differ from traditional whole transcriptome RNA sequencing?
Whole transcriptome sequencing is a relatively unbiased method for detecting fusion transcripts, enabling researchers to cast a wide net and capture surprising findings. However, this approach comes at a cost: not only is whole transcriptome sequencing expensive, but it often has lower sensitivity compared to targeted sequencing methods.
Targeted RNA sequencing, on the other hand, is distinct in that it does not require sequencing resources to be distributed across the entire transcriptome, allowing researchers to focus on specific transcripts for deep sequencing. If well-designed, targeted methods can greatly enhance detection sensitivity while reliably identifying rare fusion transcripts.
2. What are some real-world applications of targeted RNA sequencing in drug development and biological research?
Targeted RNA Sequencing is widely used in fields such as cancer research, neuroscience, plant biology, microbiology, and more. Applications include target identification for drug screening, studying disease mechanisms, and analyzing the effects of environmental stress on gene expression.
3. How does targeted RNA sequencing enhance the diagnostic capability of fusion genes?
Targeted RNA sequencing significantly amplifies the diagnostic prowess concerning fusion genes by employing biotinylated oligonucleotide probes. This method enriches RNA transcripts, facilitating the thorough identification of numerous genes, notably those that are sporadic or exhibit low expression levels. Moreover, this strategy adeptly discerns recognized fusion genes while shedding light on hitherto undisclosed fusion gene companionships.
Targeted RNA Seq Case Studies
Diagnosis of fusion genes using targeted RNA sequencing
Journal: Nature communications
Impact factor: 17.694
Published: 27 March 2019
Abstract
Fusion genes formed by chromosomal rearrangements are critical in cancer, contributing to about 20% of cases with varied prevalence across cancer types. Rapid and accurate detection is crucial for precise diagnosis and treatment. Current methods like FISH and RT-PCR, while sensitive, often miss novel fusion partners and complex rearrangements, leading to misdiagnosis in hematological cancers. RNA sequencing offers comprehensive surveillance but struggles with sensitivity for lowly expressed fusion genes. Targeted RNA-seq, using biotinylated probes, enhances detection of rare transcripts and shows promise for precise fusion gene diagnostics in solid tumors and lung cancer.
Materials & Methods
Sample Preparation
Peripheral blood samples
RNA extraction
Sequencing
Library construction
cDNA capture
Illumina HiSeq 2500 v4.0 platform
RT-PCR and Sanger sequencing
Fusion detection
In-gene coverage change
Transcriptome assembly
Immune receptor analysis
Results
Targeted RNA-seq was validated for fusion gene diagnosis in cell lines and patient tumor samples. It successfully identified known fusion genes like ROS1 and ALK in lung cancer biopsies, determining both gene partners and fusion junctions. In a broader clinical cohort of 72 samples, including solid tumors and hematological malignancies, targeted RNAseq detected fusion genes in 76% of cases, outperforming traditional methods. Reproducibility was high across replicate samples, showcasing its reliability for clinical use. The method also uncovered novel fusion genes and isoforms, highlighting its potential for guiding personalized cancer treatment.
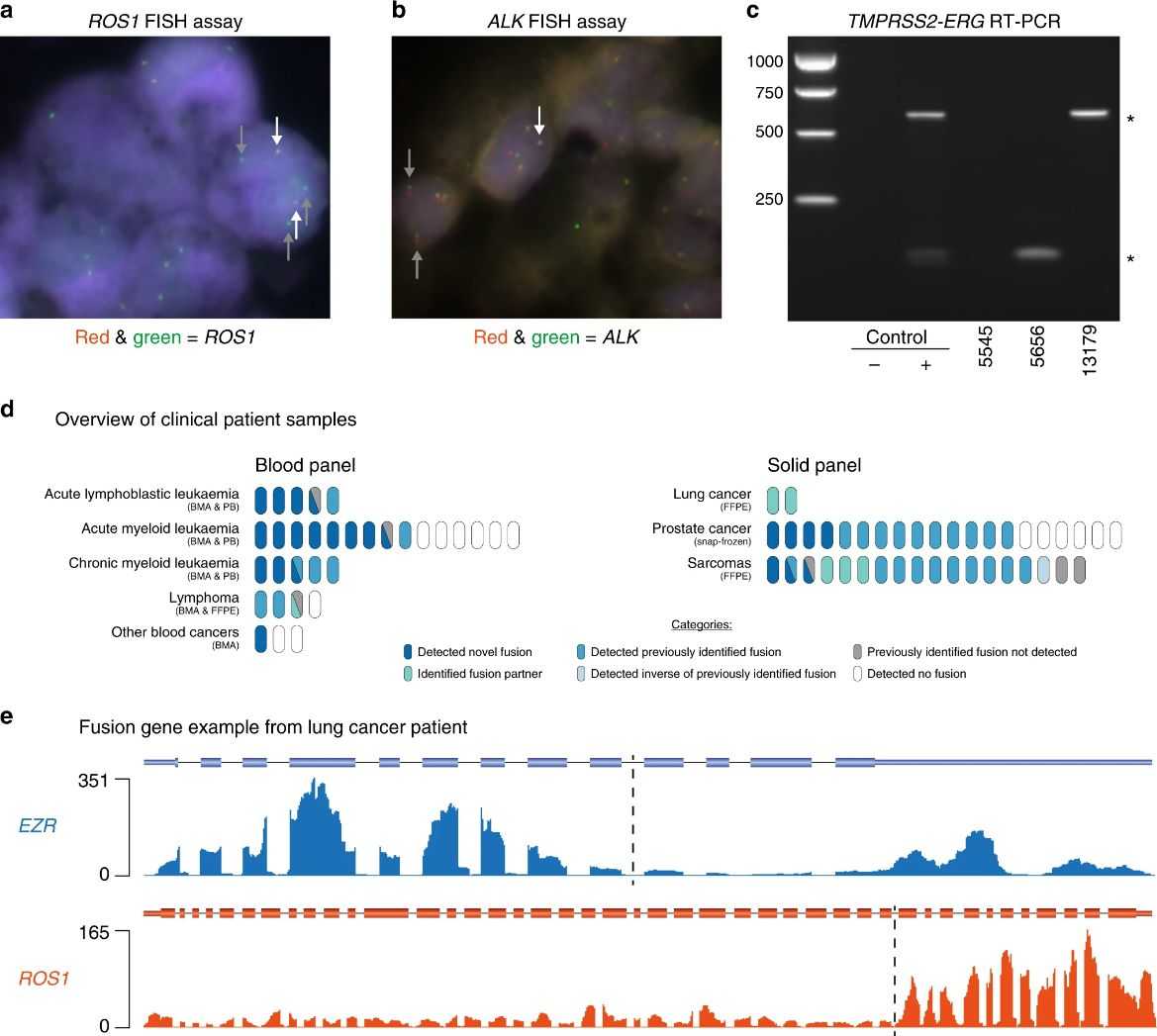 Fig 1. Fusion identification in clinical cohort samples.
Fig 1. Fusion identification in clinical cohort samples.
Targeted RNAseq not only identifies fusion genes but also quantifies gene expression across captured genes. It reveals distinct expression patterns, such as high expression of fusion genes like EZR-ROS1, and detects cases where promoter fusions drive gene expression changes, as seen with ACSL3-ETV1. Despite its sensitivity, targeted RNAseq may miss cases of promoter fusions, like in one sarcoma sample with elevated ROS1 expression. Additionally, it provides insights into marker gene expression, such as GATA2 in AML and CML, which correlates with prognosis.
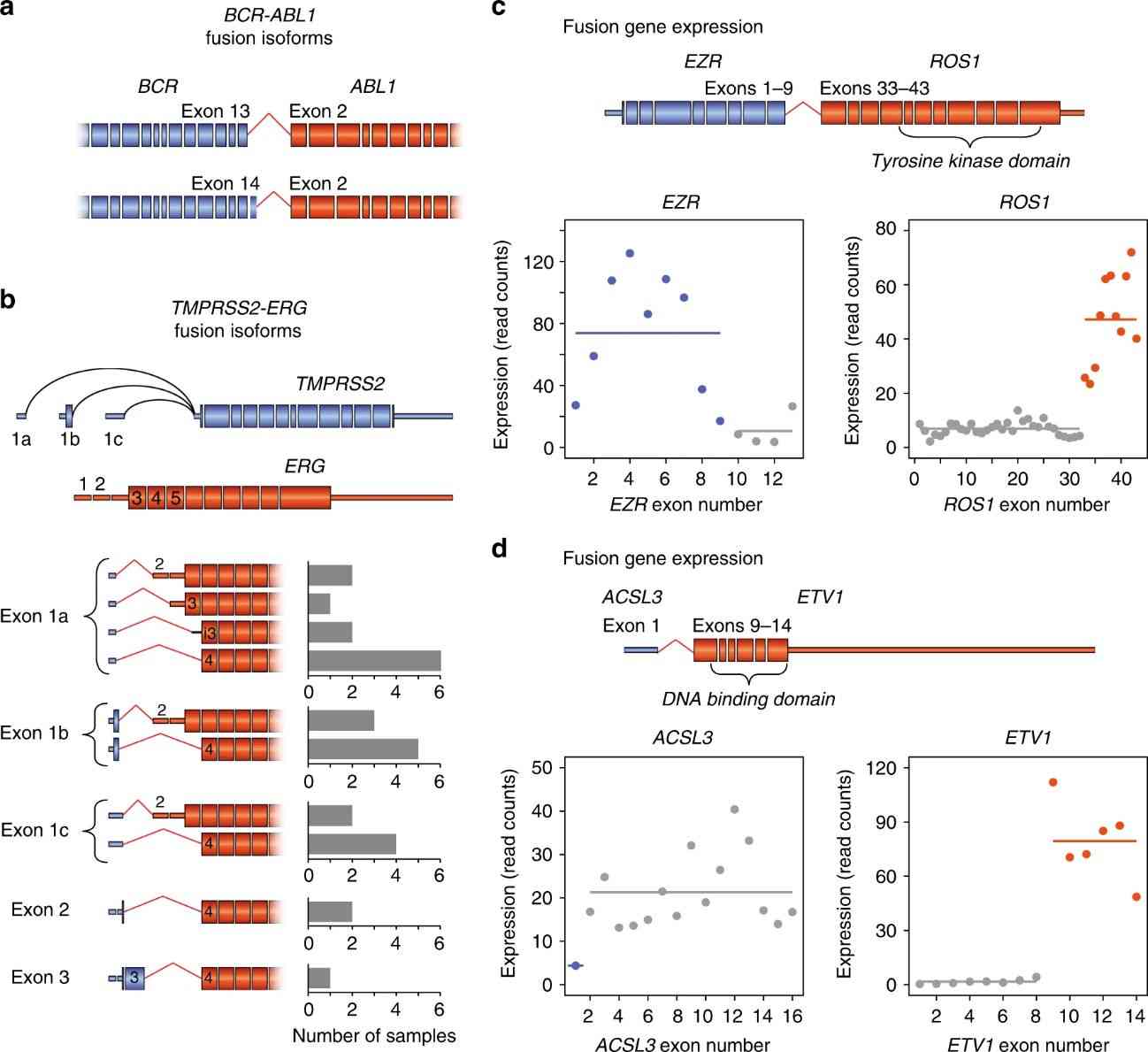 Fig 2. Fusion junction diversity and gene expression.
Fig 2. Fusion junction diversity and gene expression.
The authors' blood panel targeted V, J, and C exons at IG/TCR receptor loci to identify fusion genes like IGH-MYC and IGH-BCL6 in lymphoma patients. These probes also captured RNA transcripts from these loci, enabling profiling of the immune repertoire. Cell line analysis revealed dominant clonotypes consistent with clonal populations, while patient samples showed diverse immune receptor clonotypes, particularly in B-cell maturation in bone marrow, with notable expansions possibly indicating malignant clonal populations in some cases.
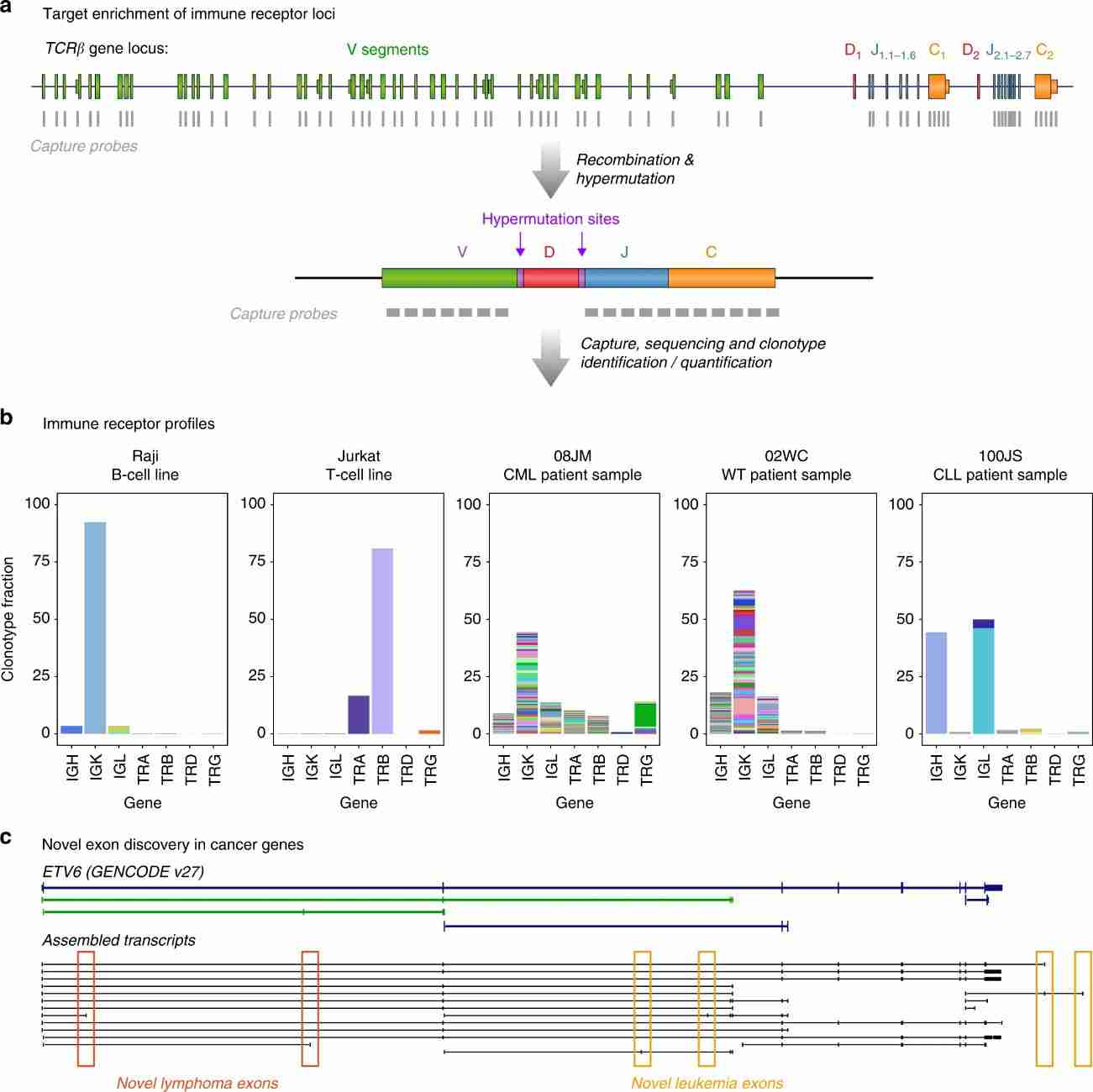 Fig 3. Novel findings in transcriptomic analysis.
Fig 3. Novel findings in transcriptomic analysis.
Conclusion
In summary, chromosomal translocations producing fusion genes are pivotal in cancer, necessitating precise diagnosis for effective treatment. Unlike traditional methods, targeted RNA-seq offers comprehensive detection of known and novel fusion genes across hundreds of genes, facilitating faster and more accurate diagnosis. Despite its high-throughput capability, targeted RNA-seq requires meticulous bioinformatic supervision to mitigate false positives. Its ability to resolve complex rearrangements and detect alternative isoforms enhances prognostic insights and treatment planning, while supplementary analyses like gene expression profiling and immune receptor clonotyping further refine diagnostic precision. Ultimately, targeted RNA-seq is poised to transform fusion gene diagnostics, offering broader clinical utility and deeper insights into cancer biology.
Reference
- Heyer EE, Deveson IW, Wooi D, et al. Diagnosis of fusion genes using targeted RNA sequencing. Nature communications. 2019, 27;10(1):1-2.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Chaperone-Mediated Autophagy Controls Proteomic and Transcriptomic Pathways to Maintain Glioma Stem Cell Activity
Journal: Cancer research
Year: 2022
Circular DNA tumor viruses make circular RNAs
Journal: Proceedings of the National Academy of Sciences
Year: 2018
Repeated immunization with ATRA-containing liposomal adjuvant transdifferentiates Th17 cells to a Tr1-like phenotype
Journal: Journal of Autoimmunity
Year: 2024
Role of the histone variant H2A.Z.1 in memory, transcription, and alternative splicing is mediated by lysine modification
Journal: Neuropsychopharmacology
Year: 2024
FAK loss reduces BRAFV600E-induced ERK phosphorylation to promote intestinal stemness and cecal tumor formation
Journal: Elife
Year: 2023
Identification of circular RNAs regulating cardiomyocyte proliferation in neonatal pig hearts
Journal: JCI insight
Year: 2024
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
