Antibody Screening Sequencing (Phage Display Library Screening)
CD Genomics is committed to offering advanced next generation sequencing (NGS) strategy to help researchers screening the phage display libraries in a high-throughput manner, enabling you to preserve the diversity of antibody libraries and quickly select the plurality of related antibodies as well as rare phages in a large population that can be recovered and tested for activity.
What is a Phage Display Library
A phage display library is a tool used in molecular biology that consists of a vast collection of bacteriophages, which are viruses that specifically infect bacteria. Each phage in the library is engineered to present a unique peptide or protein fragment on its surface. By displaying these peptides or proteins, researchers can systematically test which ones bind to a specific target molecule, such as a protein or antigen of interest. This process allows scientists to identify and isolate those with high affinity for the target, making it a powerful technique for discovering new antibodies, developing diagnostics, and exploring protein interactions.
Introduction to Phage Display Library Screening
Phage display is a powerful and widely utilized laboratory technique employed to identify peptides that bind to specific targets. This method is fundamentally based on a library consisting of millions, or even billions, of phage particles, each expressing a diverse array of exogenous peptides fused to phage coat proteins. To isolate high-affinity and high-specificity binders, multiple rounds of selection, also known as biopanning, are conducted. This process narrows down the highly diverse library to a few lead compounds and requires amplification to enrich phage clones that present target-binding peptides. Phage display is particularly crucial in the discovery and engineering of antibodies, as antibody fragments (such as scFv or Fab) can be easily expressed and displayed on phage surfaces.
The crux of phage display is the analysis of peptide sequences present in the library. Traditional Sanger sequencing necessitates the isolation of DNA from individual phage clones, a labor-intensive and small-scale methodology reliant on repeated amplification. This approach is limited by its inability to process more than a hundred library clones and may introduce unpredictable biases toward certain antibody sequences in the bacteria.
Illumina deep sequencing offers substantial advantages for analyzing phage display screens, especially for comprehensive sequence collections. It allows for fewer rounds of selection and can potentially identify binders with a single round, mitigating issues of unnecessary biases and diversity collapse introduced during amplification. Thus, it facilitates the discovery of ligands that may have been previously overlooked or underrepresented in phage display studies. Additionally, NGS platforms can characterize up to 10^6-10^8 sequences in a single run, yielding a more representative and meaningful content coverage. The strategy for sequencing phage libraries using NGS is illustrated in Figure 1.
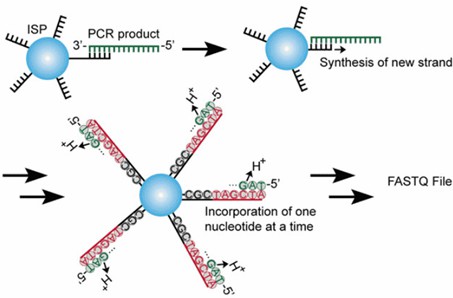 Figure 1. Workflow of antibody screening from phage display library by NGS.
Figure 1. Workflow of antibody screening from phage display library by NGS.
Our specialists can guide you to design the PCR primers which contain sequences flanking the variable region, adapter and barcodes for the enrichment of specific candidates in the most effective ways. In addition, we have developed a bioinformatics analysis pipeline that is well suited for antibody sequence analysis to enhance the discovery.
Advantages of Our Phage Display Library Screening Service
- Considerable accuracy. The ultra-deep sequencing gives high coverage and high-quality data.
- Quick selection. The selection round can be minimized to one round.
- Characterization of the affinity and stability of antibodies. Extensive data provide deeper insight.
- Cost-effective. Multiple complex samples can be tested in parallel, enable reduced cost per sample.
- Comprehensive support. We have a team consists of specialists in the field of antibody and NGS.
Applications of Phage Display Library Screening
- Development of Diagnostic Reagents: Identifying scFv sequences for the development of sensitive and specific diagnostic assays.
- Immune Cell Therapy Product Development: Constructing CAR-T and CAR-NK vectors for targeted immunotherapy in diseases like cancer.
- Disease Treatment: Utilizing scFvs in the treatment of cancers, autoimmune diseases, and age-related macular degeneration.
Phage Display Library Screening Workflow

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Phage Display Library Screening for your writing (customization)
CD Genomics provides comprehensive Antibody Screening Sequencing (Phage Display Library Screening) services designed to support the discovery and characterization of specific antibodies. Our advanced platforms enable the efficient construction of phage display libraries and subsequent high-throughput sequencing to identify and validate antibody candidates. We offer end-to-end solutions, including phage library construction, antibody screening, and sequence analysis. With our state-of-the-art technologies, we ensure accurate and rapid identification of antibody sequences, helping you to streamline your research and development processes. For any specific needs or further inquiries, our team is ready to assist.
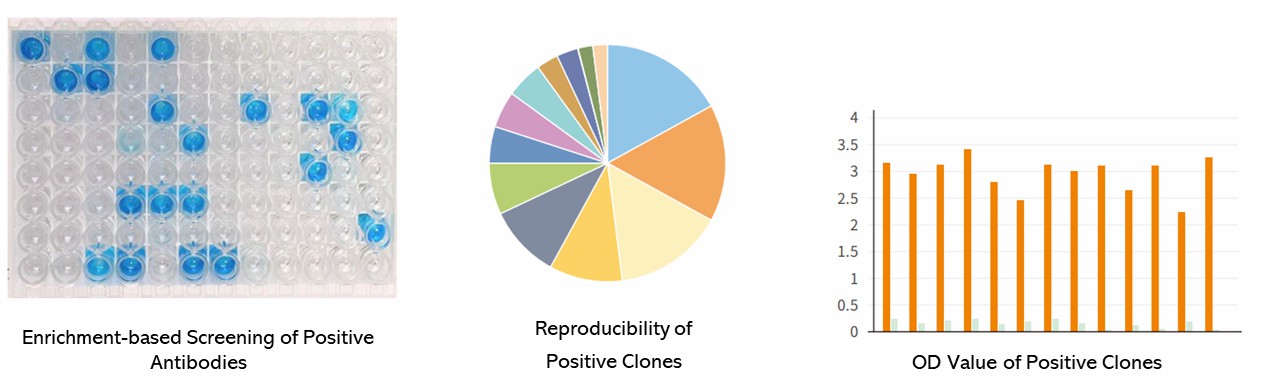
Demo Results
Partial results are shown below:
Antibody Screening Seq FAQs
1. What are the differences between preparing single-chain antibodies using phage display libraries and traditional antibody preparation methods?
When contrasting the methodologies of phage display library screening for single-chain variable fragment (scFv) antibodies with the traditional hybridoma technique for antibody production, several key distinctions become apparent.
(1) Phage Display Library Screening:
Advantages:
- Efficiency in Mimicking Immune Responses: The phage display technique can simulate the antibody production process of the animal immune system, offering several unique benefits that are difficult for hybridoma technology to match. Notably, phage display does not require immunization. In theory, a library size ranging from 10^8 to 10^10 can encompass the entire range of possible antibodies.
- Direct Screening and Rapid Expression: Specific antibodies can be directly screened from non-immune animal antibody libraries using antigens. The identified positive clones can then be recombined into plasmid expression vectors, allowing functional antibody molecules to be expressed directly in Escherichia coli or eukaryotic cells. The process is relatively swift, typically taking 16-20 weeks to construct and screen a natural antibody library.
Disadvantages:
- Affinity Considerations: The affinity of scFv antibodies obtained through phage display is often lower or comparable to that of full-length antibodies derived via traditional hybridoma techniques. However, this drawback can be mitigated if it does not impede the recognition of endogenous samples or the preservation of secondary and tertiary structures.
(2) Traditional Hybridoma-Based Antibody Preparation:
Advantages:
- Affinity Maturation and Stability: Following multiple immunizations, animals generate mature affinity antibodies. B lymphocytes from these animals are then fused and subjected to repeated subclonal screenings to obtain hybridoma cell lines that can stably secrete specific antibodies. This method produces natural full-length antibodies with higher affinity compared to the scFv antibodies from phage display libraries.
- Broad Applicability and Consistency: The resultant antibodies from hybridoma techniques are broadly applicable across varied experimental types, and they exhibit minimal batch-to-batch variation.
Disadvantages:
- Extended Timeframe: The process duration is substantially longer. From antigen preparation to the acquisition of single-chain antibodies, at least five months are required.
2. Why is there cross-reactivity between phage supernatant from positive clones and tag ELISA detection?
The phage titer in the supernatant of positive clones is typically very high, as evidenced by previous positive tests. Even when diluted by 10 or 100 times, the OD (optical density) readings can still reach around 2.0. This suggests that a small amount of non-specific adsorption is quite normal.
During the biopanning process, the phages that bind to the target are selected, but only a few clones are identified as positive after ELISA screening. The phenomenon of cross-reactivity in the ELISA test can be attributed to several factors. Most notably, it occurs within the precise balance of antigen and antibody concentrations. For indirect ELISA, especially, both the antigen and antibody need to be at appropriate concentrations to minimize non-specific binding. High concentrations of either component can exacerbate non-specific interactions, leading to what appears as cross-reactivity.
In summary, the high phage titer, along with suboptimal antigen-antibody concentration balances, contribute to the non-specific adsorption and observed cross-reactivity in tag ELISA detection.
Antibody Screening Seq Case Studies
Selection and Characterization of YKL-40-Targeting Monoclonal Antibodies from Human Synthetic Fab Phage Display Libraries
Journal: Nature biomedical engineering
Impact factor: 29.2
Published: 09 October 2023
Background
Massively parallel assays and NGS enhance throughput and speed in biomedical research, crucial for discovering antibodies and other biomolecules. However, traditional selection methods and NGS alone can suffer from biases and inefficiencies. The "deep screening" method integrates NGS with functional screening to efficiently identify high-affinity antibodies from large libraries, accelerating discovery from months to a few days and improving performance through machine learning.
Materials & Methods
Sample Preparation
- Human
- DNA extraction
- RNA synthesis
Sequencing
- NGS
- Ribosome display
- Library preparation
- Deep screening
- Image processing
- PCA analysis
- Assembly
- Statistical analyses
Results
In this proof-of-concept study, deep screening was applied to a yeast display library for rapid discovery of high-affinity binders. Following pre-selection steps, the method combined extensive sequencing and functional screening to identify 47 and 53 unique candidates from MACS and FACS selections, respectively. The technique demonstrated effectiveness in correlating high fluorescence signals with low nanomolar binding affinities and revealed deep screening's advantage in providing a comprehensive view of library performance, overcoming some biases inherent in traditional selection methods.
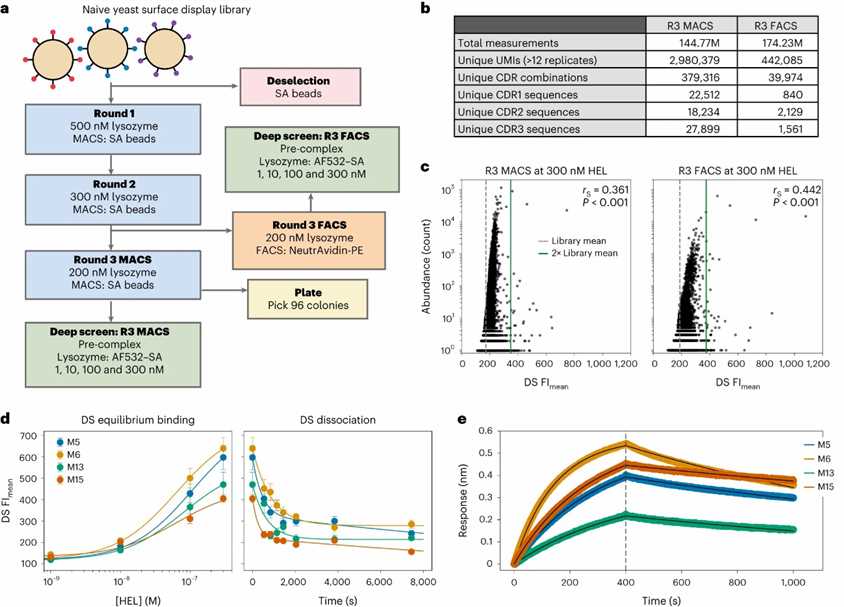 Fig. 1: Deep screening of a yeast-display-pre-selected library.
Fig. 1: Deep screening of a yeast-display-pre-selected library.
The study demonstrates that deep screening can efficiently discover high-affinity antibodies directly from an unselected scFv library. By applying this approach to a library derived from a lead candidate, IL70001, the researchers identified multiple high-picomolar-affinity Fabs against human interleukin-7, achieving up to 2,300-fold improvement over the parent antibody. Deep screening bypasses pre-selection biases, providing a rapid and direct route to high-affinity antibodies, with promising properties for further development.
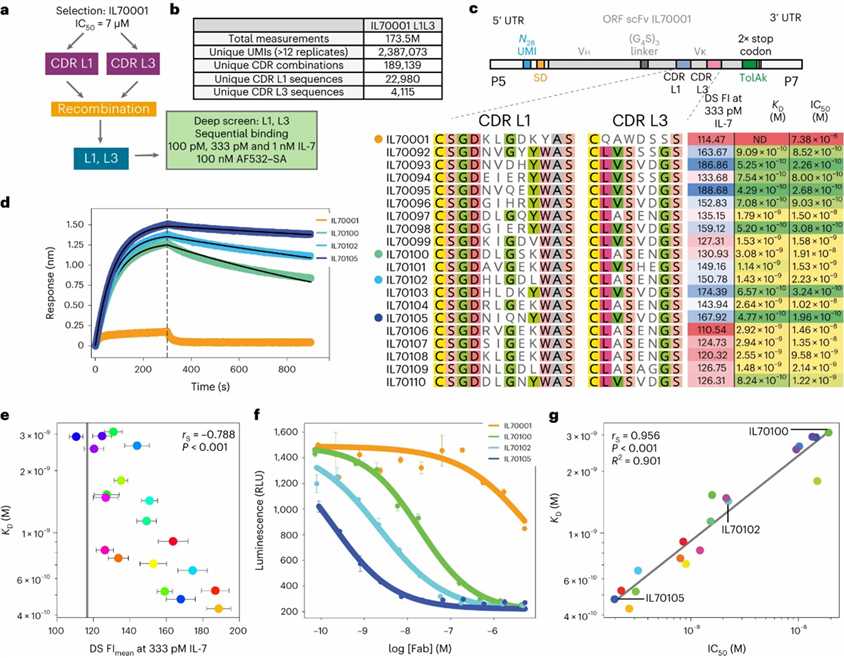 Fig. 2: Deep screening of an unselected scFv library.
Fig. 2: Deep screening of an unselected scFv library.
Conclusion
Deep screening is a high-throughput method using advanced sequencing to directly find high-affinity antibodies from unselected libraries. It improves affinity and potency by up to 10,000-fold in just three days, providing detailed, digital data on antibody interactions and supporting efficient discovery of rare, high-affinity clones.
Reference
- Porebski B T, Balmforth M, Browne G, et al. Rapid discovery of high-affinity antibodies via massively parallel sequencing, ribosome display and affinity screening. Nature biomedical engineering, 2024, 8(3): 214-232.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
