Lentiviral/Retroviral Integration Sites Analysis
Introduction to Lentiviral/Retroviral Integration Sites Analysis
Lentiviral vectors are widely recognized as the predominant gene delivery systems for achieving stable genetic modification of cells. These vectors integrate into the host genome to facilitate the expression of transgenes. Although the integration process of the lentiviral RNA genome—first reverse-transcribed into DNA—is ostensibly random, existing literature indicates the presence of integration hotspots. Given the potential for lentiviral integration to disrupt cellular function, particularly through proximity to oncogenes or critical genes, it is imperative to identify the precise integration sites post-infection. The selection of integration sites significantly influences both transgene expression and host cell phenotype. Consequently, integration site analysis is an essential tool for evaluating the safety of these vectors and monitoring the clonal dynamics of modified cells in vivo.
Our laboratory has developed an advanced suite of methodologies for comprehensive viral integration site analysis. By integrating PCR and sequencing-based techniques with proprietary bioinformatics pipelines, we provide unparalleled precision and sensitivity in identifying integration site positions and quantifying viral vector frequencies. Our end-to-end service encompasses PCR amplification, library construction, paired-end (PE) sequencing, and rigorous bioinformatics analysis. We are committed to delivering the highest quality data and reliable analytical reports, ensuring robust and reproducible results for our clients.
Methods for Integrative Site Analysis
The initial exploration of viral integration sites employed Southern blotting and genomic libraries to characterize integration site features. Over time, polymerase chain reaction (PCR) techniques have supplanted Southern blotting due to their simplicity and accuracy. Depending on the PCR methodology used, viral integration site analysis can be categorized into Reverse PCR, Ligated Mediated PCR (LM-PCR), and Linear Amplification Mediated PCR (LAM-PCR).
- Reverse PCR
Reverse PCR involves the circularization of DNA fragments following enzymatic digestion. Specific primers are then designed to amplify sequences flanking the viral integration site. However, the low efficiency of circularization with ligase limits its widespread application.
- LM-PCR
In LM-PCR, DNA is fragmented enzymatically, and adaptors are ligated to both ends of the fragments. Primers that recognize both viral DNA and the fixed adaptor sequences are utilized to amplify integration sites, thereby unlocking their positional information. LM-PCR stands out for its straightforward principle and ease of operation, retaining its relevance from inception to the present day.
- LAM-PCR
LAM-PCR begins with the linear amplification of DNA, producing single-stranded products. These single-stranded molecules are then subjected to further amplification via LM-PCR. The introduction of linear amplification significantly enhances both the sensitivity and specificity of LAM-PCR, making it superior to Reverse PCR and LM-PCR and leading to its broad adoption.
- Next-Generation Sequencing for Lentiviral Integration
Currently, high-depth sequencing, or next-generation sequencing (NGS), represents the principal platform for detecting lentiviral integration sites. The most refined and widely utilized method for creating enriched libraries for integration site analysis in industrial applications and clinical research involves mechanical fragmentation and adaptor-based amplicon library preparation (such as LM-PCR techniques like INSPIIRED). This methodology has been applied extensively in the safety evaluation of various CAR-T19 clinical projects as well as in retrospective pharmacodynamic analyses related to CAR-T cell proliferation.
Advantages of Our Lentiviral/Retroviral Integration Sites Analysis Service
- Mature integration site analysis techniques.
- Multiplex samples for cost-effective results.
- Effective workflow and fast turnaround time.
- Qualitative and quantitative analysis.
- Comprehensive bioinformatics analysis.
- Multiple approaches to meet different goals.
Applications of Lentiviral/Retroviral Integration Sites Analysis
Safety Assessment:
- Avoiding Insertional Mutagenesis
- Monitoring Long-Term Effects
Virology Research:
- Understanding Viral Life Cycle
- Developing Vaccines and Antiviral Drugs
Optimizing Transgenic Animal Models
- Enhancing Stability and Expression
- Reducing Adverse Effects
Regenerative Medicine and Cell Research
- Improving Research Efficiency
- Tracking Cell Fate
Basic Biological Research
- Exploring Genome Structure and Function
- Investigating Genomic Rearrangements
Lentiviral/Retroviral Integration Sites Analysis Workflow
Our highly experienced expert team executes quality management following every procedure to ensure comprehensive and accurate results. Our Lentiviral Integration Sites Analysis workflow is outlined below, including library prep, sequencing, and bioinformatics analysis.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Lentiviral/Retroviral Integration Sites Analysis for your writing (customization)
CD Genomics' lentiviral integration sites analysis service utilizes the power of Illumina Platform for integration sites detection, and has developed a low-cost high-throughput approach by using target enrichment sequencing or LM-PCR method. We are pleased to use our extensive experience and advanced platform to offer the best service and the most qualified products to satisfy each demand from our customers.
Reference
- Dawes JC, Webster P, Iadarola B, et al. LUMI-PCR: an Illumina platform ligation-mediated PCR protocol for integration site cloning, provides molecular quantitation of integration sites. Mob DNA. 2020; 11:7.
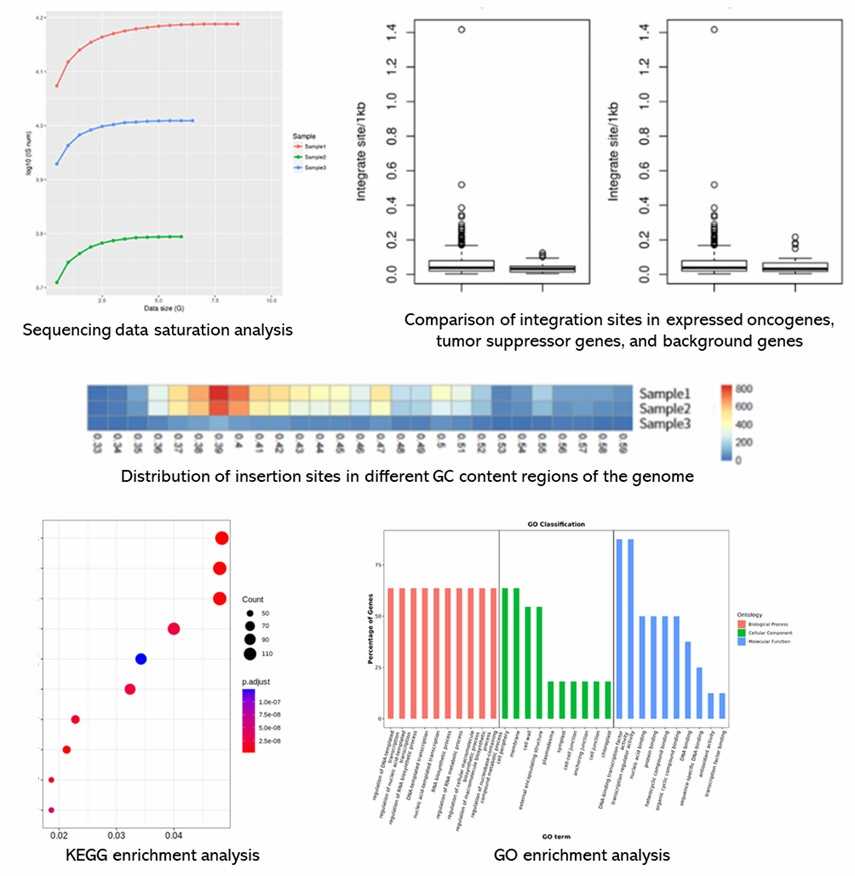
Demo Results
Partial results are shown below:
Lentiviral/Retroviral Integration Sites Analysis FAQs
1. What is Lentiviral/Retroviral Integration Site Analysis?
Lentiviral/Retroviral Integration Site Analysis is a sophisticated technique used to examine how lentivirus and retrovirus genomes integrate into the host cell genome. By identifying and mapping these integration sites, researchers gain crucial insights into the mechanisms of viral integration, assess the safety of the vectors, and monitor the in vivo behavior of transgenic cells.
2. Why is Integration Site Analysis Necessary?
Integration site analysis is vital for evaluating the safety of viral vectors. It ensures that these vectors do not insert into critical regions of the host genome, which could potentially trigger oncogenesis or other genetic disorders. Additionally, this analysis aids researchers in understanding the preferences and mechanisms of viral integration, thereby providing essential support for developing safer and more effective vector-based approaches.
3. What are the Basic Steps of Integration Site Analysis?
The fundamental procedure for lentiviral/retroviral integration site analysis includes the following steps:
- Infection of Target Cells: Infect the target cells with the viral vector.
- Isolation of Genomic DNA: Extract the genomic DNA from the infected cells.
- Restriction Enzyme Digestion: Digest the isolated genomic DNA using restriction endonucleases.
- Ligation of Adapter Sequences: Ligate adapter sequences to the digested DNA fragments.
- PCR Amplification: Perform PCR amplification to enrich for DNA fragments containing viral integration sites.
- Library Construction: Construct sequencing libraries from the enriched PCR products.
- High-Throughput Sequencing: Conduct high-throughput sequencing, such as Illumina sequencing.
- Data Analysis and Integration Site Identification: Analyze the sequencing data to identify and characterize the viral integration sites.
4. What Tools and Techniques are Required?
Integration site analysis typically involves the following tools and techniques:
- Restriction Endonucleases: Enzymes used to digest genomic DNA at specific sequences.
- PCR Amplification: Techniques for selective amplification of DNA sequences containing viral integration sites.
- Adapter Ligation: The method of attaching adapter sequences to DNA fragments to facilitate sequencing.
- High-Throughput Sequencing: Platforms such as Illumina sequencing for generating extensive sequence data.
- Bioinformatics Analysis Tools: Software and algorithms for sequence alignment (e.g., BLAST, Bowtie) and data analysis (e.g., R, Python) to elucidate integration site locations and their characteristics.
Lentiviral/Retroviral Integration Sites Analysis Case Studies
Genome-wide integration site detection using Cas9 enriched amplification-free long-range sequencing
Journal: Nucleic acids research
Impact factor: 8.278
Published: 22 February 2021
Background
Gene delivery vectors, particularly lentiviruses, are used to insert genetic material into cells. Advances in vector technology and sequencing methods have improved their safety and precision. New techniques like Nanopore MinION sequencing enable accurate mapping of vector integration sites, enhancing our understanding of their effects.
Materials & Methods
Sample Preparation
- Cells
- DNA extraction
Methods
- ddPCR
- Sequencing
- MinION platform
- Quality filtering
- Size selection
- Initial alignment
- Mapping to reference genome
Results
Analysis of integration sites reveals that lentiviral vectors integrate randomly across different cell models and species. Most integrations occur in gene regions, with a preference for introns over exons, but show no significant pattern or overlap between different vector types or cell lines. This randomness indicates that lentiviral vectors do not target specific genomic regions, suggesting a broad distribution without bias towards oncogenes.
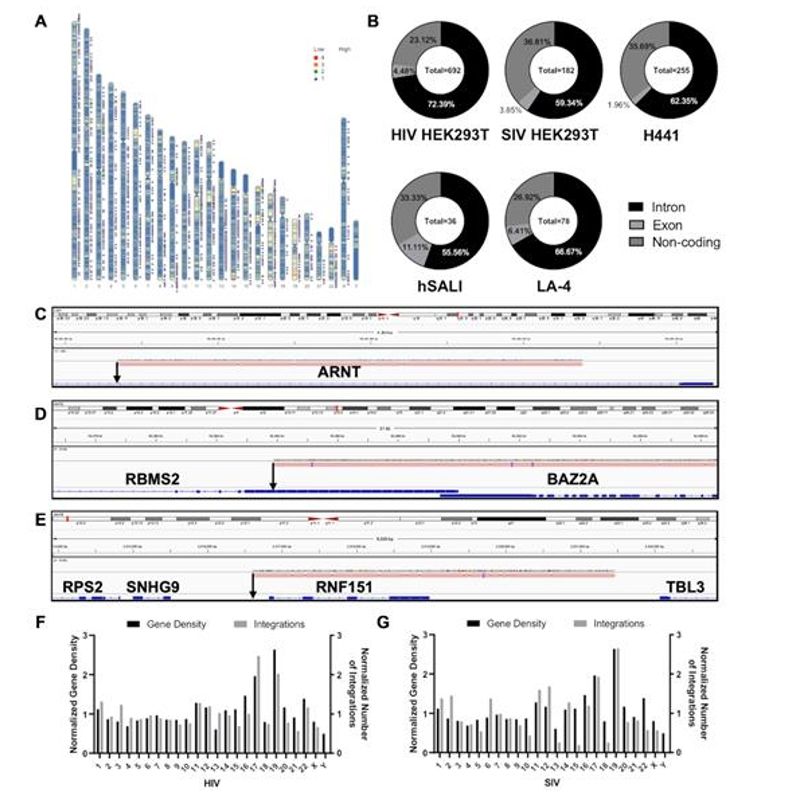 Figure 1. Positional Analysis of IS Identified.
Figure 1. Positional Analysis of IS Identified.
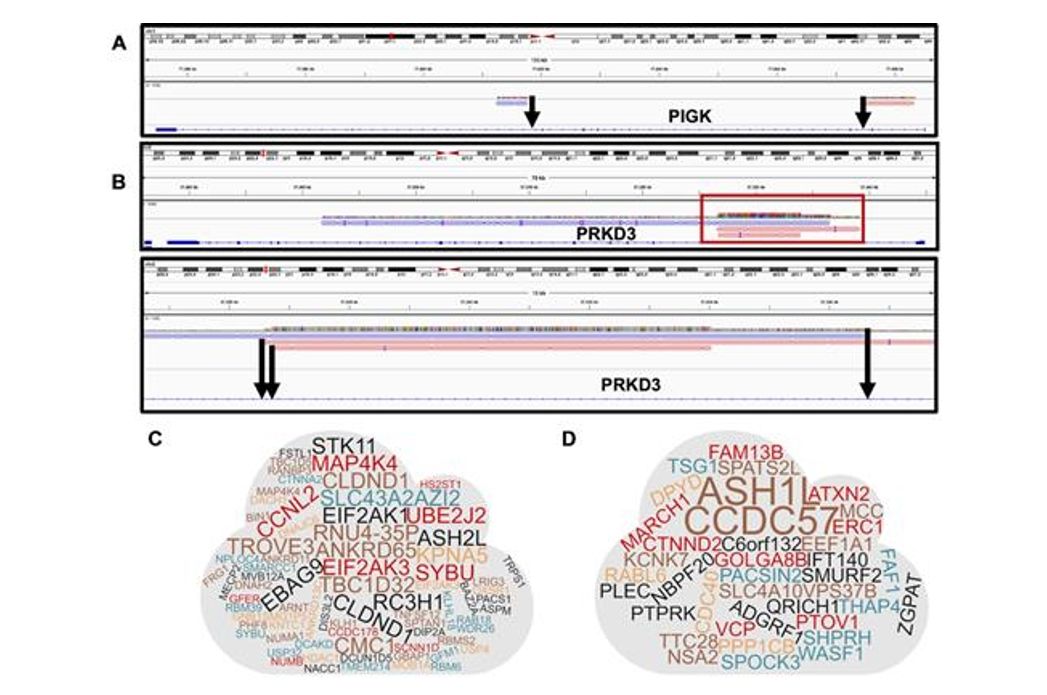 Figure 2. Identification of Multiple Integration Sites (MIS).
Figure 2. Identification of Multiple Integration Sites (MIS).
Conclusion
The method used in this study detects lentiviral integration sites without DNA amplification, employing Cas9 to attach adapters for long-read Nanopore sequencing. This approach provides faster results than traditional methods and avoids biases from PCR. It offers detailed integration site mapping with long reads, though it requires more DNA and may miss some sites due to the high input needed. The method can also be adapted for other genomic analyses and reveals additional genetic details not captured by current methods.
Reference
- van Haasteren J, Munis AM, Gill DR, et al. Genome-wide integration site detection using Cas9 enriched amplification-free long-range sequencing. Nucleic acids research. 2021, 49(3): e16-.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
