Why Choose Sanger Sequencing?
Sanger sequencing remains the gold standard for DNA analysis when accuracy is paramount. Unlike NGS, it offers unmatched single-base precision (QV ≥40), read lengths over 800 bp, and reliable results for targeted sequencing tasks.
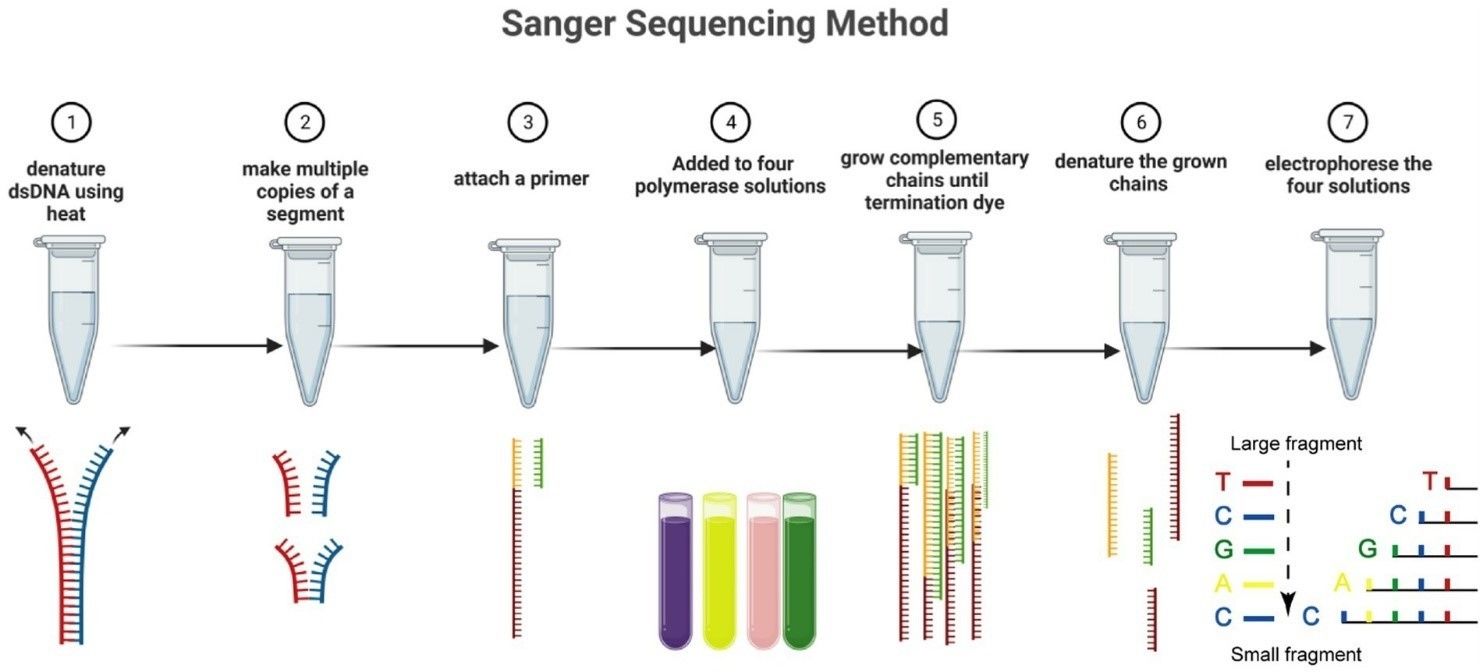 The Sanger sequencing method in seven steps. (Nafea, Aljuboori M., et al. 2024)
The Sanger sequencing method in seven steps. (Nafea, Aljuboori M., et al. 2024)
When to Use Sanger Sequencing
- Mutation verification
- Cloning and plasmid validation
- Short fragment sequencing (100 bp–1.5 kb)
- Microbial species identification
Why It Outperforms NGS in These Cases
| Feature | Sanger Sequencing | NGS |
| Single-Base Accuracy | 99.99% | ~99.9% (Often needs Sanger) |
| Ideal Fragment Length | 100 bp – 1.5 kb | Whole genome |
| Best Use Cases | Targeted, high-accuracy | Broad, high-throughput |
Where Sanger Sequencing Makes a Difference
Our high-quality Sanger sequencing services support a broad spectrum of molecular biology research, offering reliable data for applications that demand precision. Below are key areas where Sanger sequencing plays a vital role in advancing scientific discovery.

Cloning & Plasmid Validation
- Validate insert orientation and reading frame
- Confirm plasmid integrity before expression
- Detect small mutations or indels
Genotyping & Model Screening
- Identify SNPs, indels in knockout/knock-in models
- Analyze mouse tail DNA and similar genomic templates
- Determine zygosity (homo-/heterozygous)
TA Cloning & Insert Screening
- Verify directionality and frame of T-vector insertions
- High-throughput clone screening made easy
Primer Walking for Large Inserts
- Sequence long fragments >1.5 kb
- Assemble full-length gene or vector sequences
- Design and optimize primers for each read step
GC-Rich & Structured Region Sequencing
- High-fidelity reads through GC-heavy or repetitive sequences
- Specialized protocols for pseudogenes or hairpin loops
Microbial Strain Identification
- Amplify & sequence 16S/18S/ITS for species-level ID
- Suitable for food, soil, or environmental samples
Sanger Sequencing Services We Offer
| Service Type | Ideal For | Key Features |
|---|---|---|
| Standard Sanger Sequencing | PCR products, plasmids, BACs | Fast turnaround, high accuracy (QV≥40), up to 1400 bp reads |
| EZ-Seq Pre-Mixed Sequencing | Routine, high-throughput workflows | Clients mix template + primer; minimal prep; ideal for bulk projects |
| Whole Plasmid Sequencing | Full plasmid verification | Primer walking strategy; no library prep needed |
| Difficult Region Sequencing | High GC content, hairpins | Optimized protocols for challenging templates |
| Primer Walking Sequencing | Unknown or long inserts | Automated primer design; scalable read extension |
| Microbial Identification Sequencing | Bacteria, fungi, mixed samples | 16S/18S/ITS sequencing for accurate species ID |
| Rolling Circle Amplification (RCA) Sequencing | Direct colony sequencing | High-fidelity amplification; bypasses plasmid extraction |
| Custom Sequencing Solutions | Complex or unique projects | Modular workflows with PCR, synthesis, sequencing & analysis |
Tip: Not sure which service fits your sample? Contact us for a free consultation.
Complete Sanger Sequencing in 4 Steps - Service Workflow
Our efficient and user-friendly Sanger sequencing service process includes: online order submission, sample shipment, standardized laboratory operations, and rapid results delivery. Throughout the process, we provide professional support to ensure data accuracy and reliability.
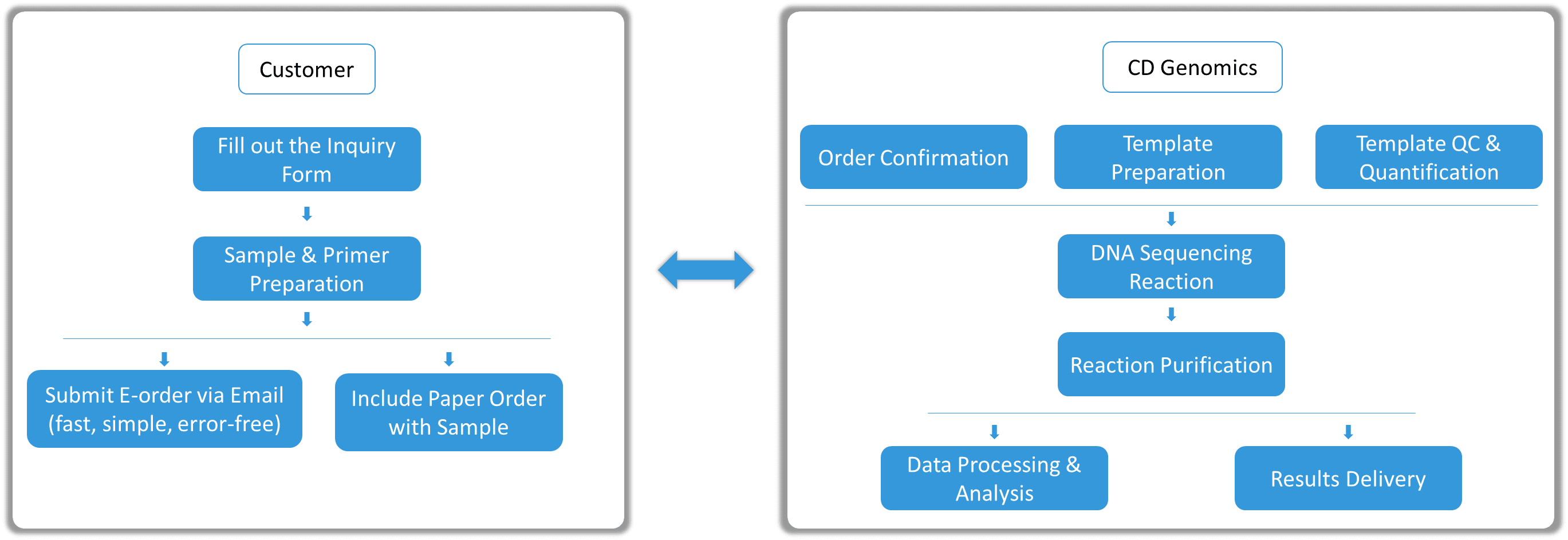
Our Sanger Sequencing Strategy
Sequencing Format
- Reaction Setup: Single template + single primer
- Throughput: 1.5 mL tubes or 96-well plate formats
- Read Length: Up to 1000 bp (high-quality region ≥800 bp)
- Data Output: .ab1 files, base-called FASTA, quality score reports
Supported Sample Types
- Purified plasmid DNA
- PCR products (clean or crude)
- High-GC or structured regions (with optimized protocols)
- Colony or liquid culture (via RCA or extraction)
Equipment
- Applied Biosystems 3730xl DNA Analyzer
- 3500 / 3500xL Genetic Analyzer
Primer Options
- Client-supplied primers
- Universal primers (e.g., M13, T7, SP6)
- Custom-designed primers upon request
Sanger Sequencing Bioinformatics Analysis
- Quality Control: Assess signal clarity from electropherogram files and trim low-quality bases.
- Accurate Base Calling: Convert chromatogram peaks into DNA sequences using trusted algorithms, with manual verification when needed.
- Sequence Alignment: Precisely align reads to reference genomes or target genes to ensure correct variant identification.
- Variant Detection & Annotation: Identify SNPs, indels, and other mutations, and annotate them using public databases.
- Contig Assembly: Merge overlapping reads into full-length sequences for complete analysis.
- Advanced Analysis: Support for mutation validation, phylogenetic studies, or custom downstream analyses based on your research goals.
CD Genomics offers end-to-end bioinformatics services to help you get the most from your Sanger sequencing data. Our pipeline ensures high accuracy and reliable interpretation:
Whether you're confirming a mutation or reconstructing a full insert, our expert team ensures your data is accurate, interpretable, and publication-ready.
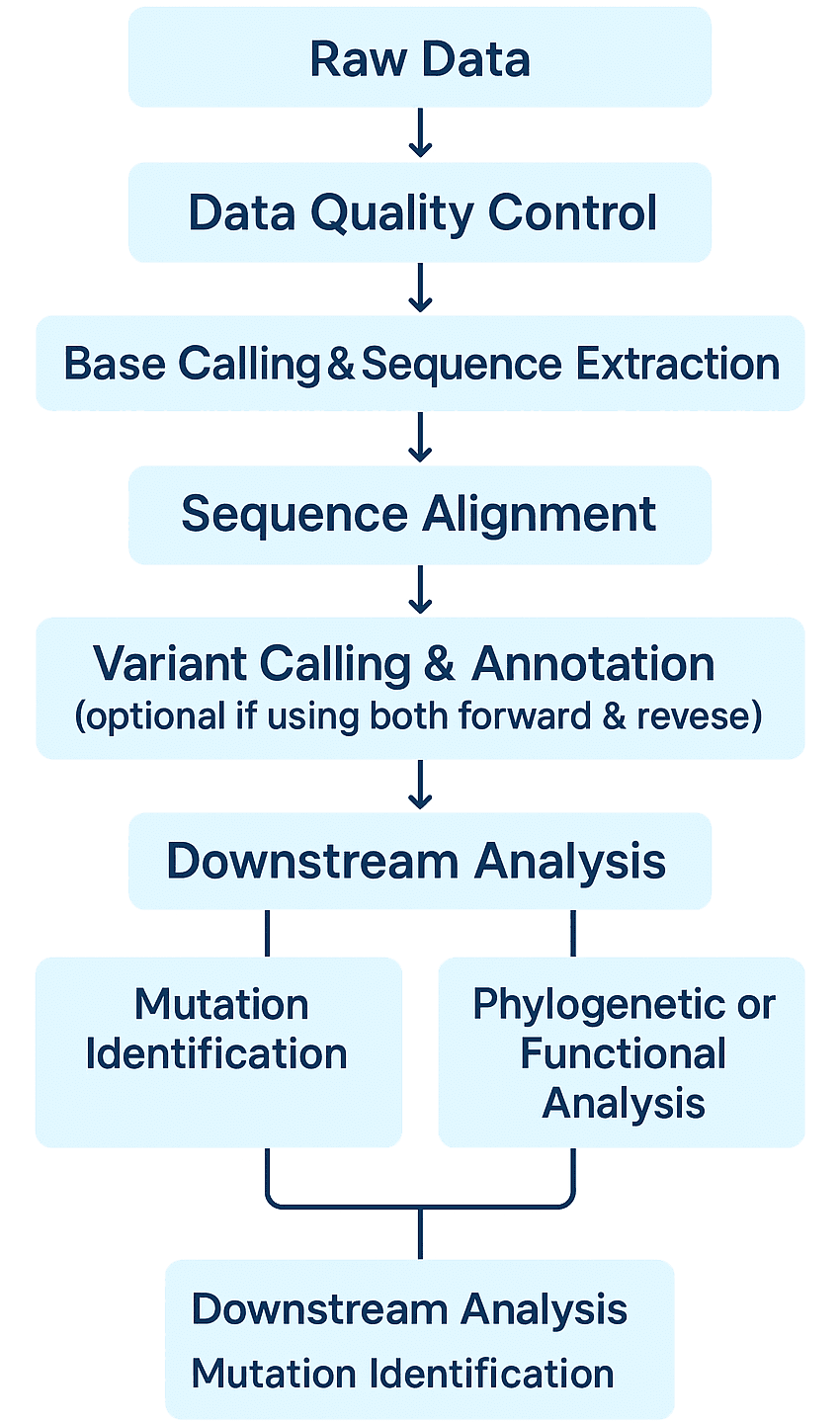
Sanger Sequencing Data Quality Assurance

At CD Genomics, we prioritize accuracy, consistency, and success—even for complex templates. Our sequencing platform is powered by Thermo Fisher's trusted systems and rigorous QC protocols, ensuring:
- Read Lengths >800 bp
Consistently long reads reduce assembly steps and boost analysis efficiency. - High Accuracy (QV40–60)
Clear signal and precise base calling for confident variant detection and alignment. - 95% Success Rate on Difficult Regions
Expert strategies for high-GC, hairpin, and repetitive sequences deliver results where others fail.
Our promise: To deliver sequencing results with stable read lengths, excellent accuracy, and high success rates, propelling your research forward with confidence.
Sanger Sequencing Sample Preparation Guide
To ensure optimal sequencing quality and reliable results, CD Genomics provides comprehensive sample preparation guidelines tailored for different template types and research requirements. Please follow the instructions below to ensure your samples meet the necessary criteria. If you have any questions or unique sample types, our technical support team is available to assist with customized solutions.
| Sample Type | Concentration Requirement | Recommended Volume | Fragment Length | Additional Notes |
|---|---|---|---|---|
| Purified Plasmid (<10 kb) | ≥ 100 ng/μL | ≥ 15 μL | Insert <10 kb | Use double-distilled water (not TE buffer) to avoid inhibition. |
| Purified Plasmid (>10 kb) | ≥ 500 ng/μL | ≥ 15 μL | Insert >10 kb | Increase concentration for long inserts to ensure complete reads. |
| PCR Product (Purified) | ≥ 20 ng/μL | ≥ 15 μL | 100 bp–3 kb | Bands must be distinct; verify via gel. Use clone sequencing for fragments <100 bp. |
| PCR Crude | Clear, single band on gel | ≥ 30 μL | 100 bp–3 kb | Indicate fragment length and purification needs; avoid non-specific bands. |
| DNA Fragments (e.g., gel purified) | ≥ 30 ng/μL | ≥ 15 μL | 100 bp–3 kb | Ensure clear and specific bands. |
| Large DNA (BACs, genomic DNA) | 50–100 ng/μL | ≥ 15 μL | >10 kb | Submit high-quality DNA; avoid shearing. |
| Bacterial Colony | Single fresh colony | — | Insert <6 kb | Indicate vector and resistance markers. For large/low-copy plasmids, send purified DNA instead. |
| Liquid Culture | Fresh culture ≥200 μL | 2–4 mL | Insert <6 kb | Indicate plasmid and resistance info. For ultra-low-copy plasmids, send purified DNA. |
🔹 Minimum submission volume: 15 μL per sample (10 μL required per reaction). For multiple reactions, increase total volume accordingly.
🔹 Note: For special samples, please contact technical support for customized solutions.
Why Choose CD Genomics Sanger Sequencing Services
- Industry Experience: Extensive genomic services experience with a global reach.
- Automated Platform: Utilizes Thermo Fisher 3730xl high-throughput sequencers across all operations.
- Leading Read Lengths: Sequencing read lengths up to 1200 bp meet most vector and product requirements.
- High Data Stability: Consistency over 99.95% in repeat sequencing.
- Quick Response: Technical support typically responds within 2 hours, offering flexible and efficient problem-solving.
- Fast Turnaround: Most samples sequenced and data delivered within 24 hours.
- Flexible Service: Supports everything from single samples to high-throughput, multi-project parallel processing.

Reference
- Nafea, Aljuboori M., et al. "Application of next-generation sequencing to identify different pathogens." Frontiers in microbiology 14 (2024): 1329330. https://doi.org/10.3389/fmicb.2023.1329330
- Crossley, Beate M., et al. "Guidelines for Sanger sequencing and molecular assay monitoring." Journal of Veterinary Diagnostic Investigation 32.6 (2020): 767-775. https://doi.org/10.1177/1040638720905833
- Dey, Pranab. "Sanger sequencing and next generation gene sequencing: Basic principles and applications in pathology." Basic and advanced laboratory techniques in histopathology and cytology. Singapore: Springer Nature Singapore, 2023. 247-261. https://doi.org/10.1007/978-981-19-6616-3_23
- Valencia, C. Alexander, et al. "Sanger sequencing principles, history, and landmarks." Next Generation Sequencing Technologies in Medical Genetics (2013): 3-11. https://doi.org/10.1007/978-1-4614-9032-6_1
- Chang, Ya-Sian, et al. "Evaluation of whole exome sequencing by targeted gene sequencing and Sanger sequencing." Clinica Chimica Acta 471 (2017): 222-232. https://doi.org/10.1016/j.cca.2017.06.015
Demo Results
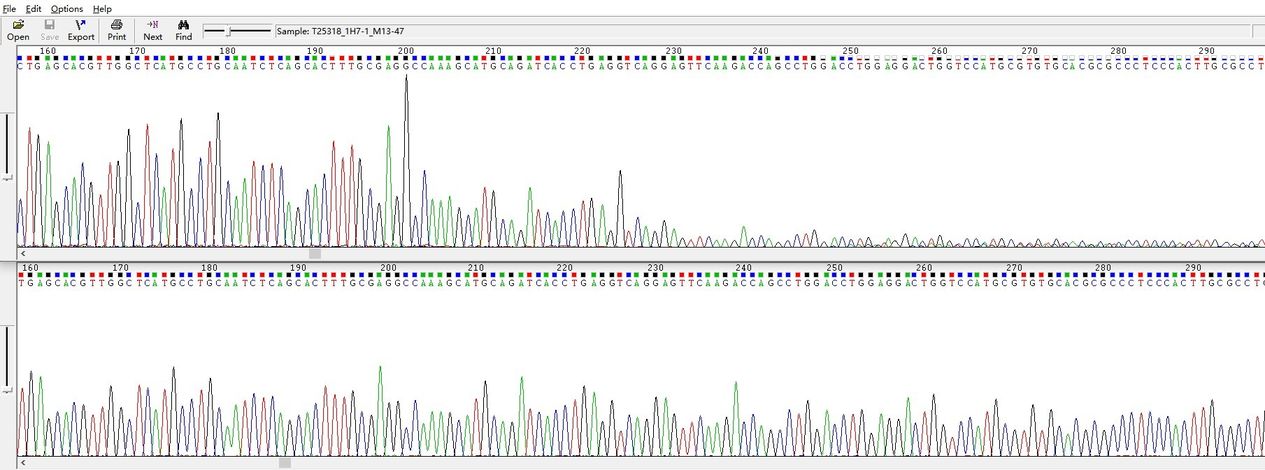
220bp Hairpin Structure
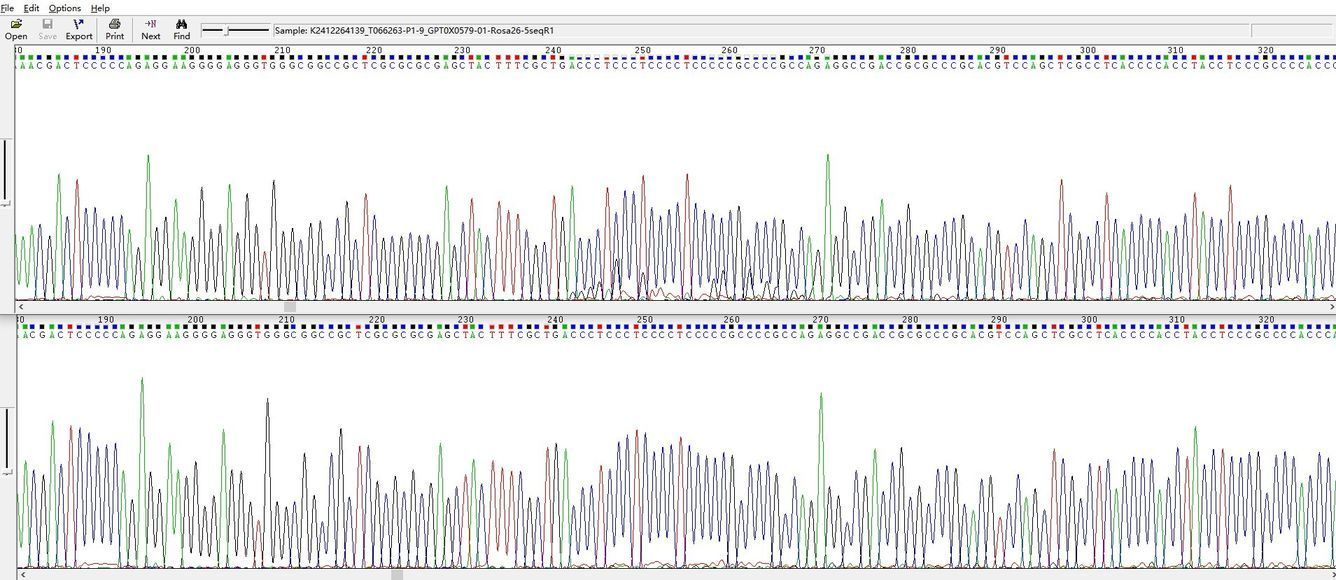
240–260bp High GC Content Structure
Sanger Seq FAQs
What solution is best for dissolving DNA sequencing samples?
We recommend using sterile distilled water. DNA sequencing depends on the polymerization reaction of Taq polymerase, and solutions with buffering salts like TE Buffer can interfere with the reaction system, reducing sequencing efficiency. Although TE Buffer is beneficial for DNA preservation, it can affect the reaction conditions, so using sterile distilled water to dissolve samples is advised.
What form is best for providing DNA sequencing samples?
We suggest clients provide bacterial cells (such as stab cultures or fresh liquid cultures) for plasmid extraction by us, as this ensures greater stability. If you provide DNA samples, ensure their purity and concentration meet standards. Especially for PCR products, perform gel purification to ensure sequencing quality. Refer to the “DNA Sequencing Sample Preparation and Considerations” for more details.
In what form should bacterial cell samples be provided for sequencing?
Sending stab cultures or fresh liquid cultures is recommended. Plate cultures are prone to damage during transport, and glycerol stocks can be easily contaminated. Stab cultures can be prepared by inoculating agar in a 1.5ml tube with a toothpick and incubating overnight at 37°C. This method is stable, convenient for transport, and can be stored at 4°C for several months.
Why are very short PCR products not suitable for direct sequencing?
Short fragments (<150 bp) are unsuitable for direct sequencing because:
- Accuracy in the 30–50 bp region following the sequencing primer is low.
- They are too short for easy purification.
- They are more susceptible to external interference, affecting result accuracy.
Thus, PCR product lengths should be at least 150 bp; otherwise, cloning prior to sequencing or redesigning primers is advisable.
What if a DNA fragment is too long to be covered in a single sequencing reaction?
Consider using bidirectional sequencing or Primer Walking strategy:
- Sequence using primers from both ends first.
- If the full length is still not covered, design new primers in the 500–700 bp region already sequenced to continue until complete.
Why are primers in Primer Walking typically designed so close to the beginning?
Primer design must ensure:
- The sequence in the region is accurate.
- The subsequent area is reliable enough for assembly.
Therefore, we often choose positions slightly ahead of the known 3’ end sequence to ensure primer effectiveness and assembly accuracy.
I have a 4 kb PCR fragment; can you help sequence it completely?
For PCR fragments longer than 3 kb, it is advisable to first clone them into a vector before sequencing. This method provides higher template stability and better sequencing results, making it easier to obtain a complete sequence.
Sanger Seq Case Studies
Customer Publication Highlight
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal:
Hypertension
Impact factor: ~10.0
Published: 10 May 2022
DOI: https://doi.org/10.1161/HYPERTENSIONAHA.121.18711
Background
A significant proportion of individuals with hypertension exhibit reduced responsiveness to certain medications. Emerging evidence suggests gut microbiota may influence drug metabolism by enzymatically degrading compounds, altering their bioavailability. This study investigated how Coprococcus comes, a gut commensal, compromises the blood pressure (BP)-lowering effects of ester-based angiotensin-converting enzyme (ACE) inhibitors through enzymatic degradation.
Project Objective
The client aimed to:
- Validate the role of Coprococcus comes in hydrolyzing ester ACE inhibitors.
- Identify microbial contributions to reduced drug efficacy using sequencing and functional assays.
CD Genomics’ Services
As a leader in Sanger sequencing, CD Genomics provided critical support:
- Strain Identification: Sanger sequencing of 16S rRNA confirmed C. comes purity after anaerobic culture.
- High-Fidelity Sequencing: Validated bacterial colony identity with >99% accuracy.
- Data Integration: Combined sequencing results with HPLC-MS and BP measurements for mechanistic insights.
Key Findings
- Gut Microbiota Modulates Drug Bioavailability:
- Depleting gut microbiota in hypertensive rats (SHR) via antibiotics enhanced oral ester ACE inhibitor efficacy (MAP reduction: −25 mmHg vs. control −15 mmHg, P < 0.001).
- Intravenous administration bypassed microbial interference, confirming gut-specific catabolism.
- Coprococcus comes Exhibits Esterase Activity:
- Sanger sequencing confirmed C. comes identity in cultured isolates.
- In vitro assays showed 68% degradation of ester ACE inhibitors via carboxylesterase (EC 3.1.1.1).
- Structural Selectivity in Drug Catabolism:
- C. comes hydrolyzed ester ACE inhibitors but spared non-ester variants, highlighting enzyme specificity.
- Demographic Disparity in Microbial Composition:
- Metagenomic analysis of human fecal samples revealed higher Coprococcus abundance in one demographic group (P < 0.05), aligning with trends in drug responsiveness.
Figures Referenced
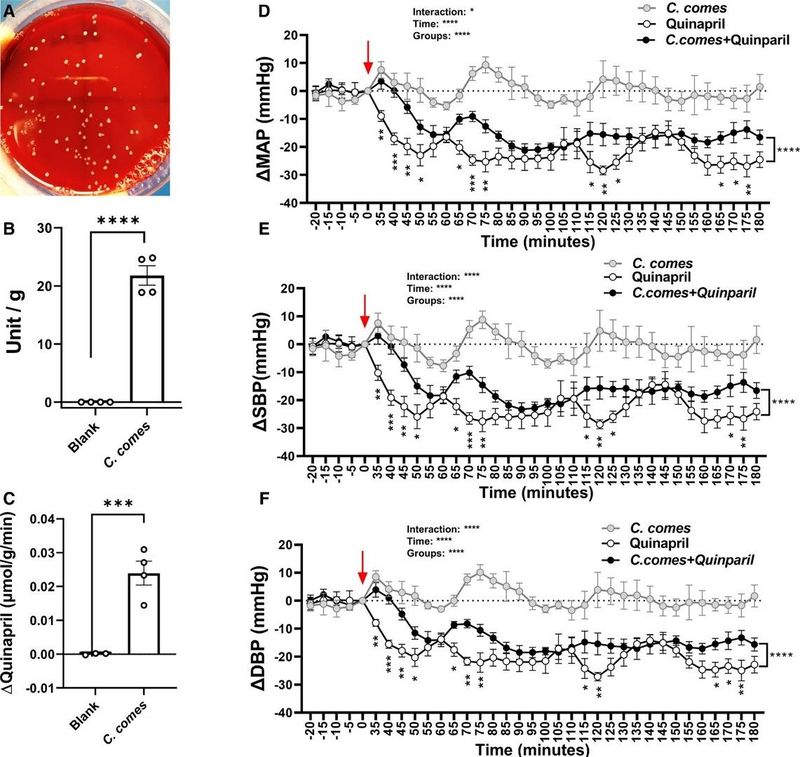 C. comes-mediated hydrolysis of ester ACE inhibitors.
C. comes-mediated hydrolysis of ester ACE inhibitors.
Reference
- Yang, Tao, et al. "Identification of a gut commensal that compromises the blood pressure-lowering effect of ester angiotensin-converting enzyme inhibitors." Hypertension 79.8 (2022): 1591-1601. https://doi.org/10.1161/HYPERTENSIONAHA.121.18711
Related Publications
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
