Genotyping
CD Genomics provides a fully flexible genotyping service for small, large, standard or customized projects, on humans and many other species. Our powerful portfolio includes arrays, reagents, instruments and bioinformatics tools that enable you to detect common and rare single nucleotide polymorphisms (SNPs), copy number variations (CNVs) and other genetic variations. CD Genomics genotyping services accommodate projects with a broad range of applications. With more than 10 years of service experience, we offer a highly trusted, flexible and competent service combined with high industry standards and scalable capacity, and we have been recognized as the premium choice of SNP genotyping.
What is Genotyping
Genotyping serves as a scientific process by which the particular alleles located at given gene sites within an organism's genome are identified and described. In the realm of biological research, the fundamental purpose of genotyping is to ascertain the types of unique alleles harboured by an organism at designated gene loci. This practice aids in unveiling the correlation between genotype and phenotype, and the influence of genetic variations on an organism's traits and phenotypic expression. Genotyping uncovers genetic disparity by comparing a DNA sequence with another sample or a reference sequence. This technique facilitates the detection of subtle alterations in a population's gene sequences, like single nucleotide polymorphisms (SNPs).

Single Nucleotide Polymorphisms (SNPs) are bi-allelic (usually) nucleotide variants that have been found throughout the genome at a frequency of about one in 1,000 bp. They can be found in coding, non-coding, and intronic regions of genomes, and they may affect transcription factor binding, gene splicing, protein folding, and many other elements at the gene and transcript level. Hence they may be responsible for the diversity among individuals and genome evolution, and are ideal markers for identifying genes associated with complex diseases. SNPs are one of the two most common sources of genetic variation (the second one are copy number variants, CNVs). SNP analysis is important and quite meaningful for studies such as disease related genetics, individualized health management, breeding, etc.
On the basis of the purposes of the study, SNP genotyping can be divided into two domains, whole genome genotyping and fine mapping. Whole genome genotyping is a platform for screening SNPs. Fine mapping here is defined as SNP genotyping analysis at a high density for selective genomic regions. Fine mapping often follows large-scale genome-wide studies to zoom into potential genes associated with the disease of interest, and requires the validation of fewer (e.g., fewer than 1,000) SNPs highly specific for each disease for a larger sample size, it should achieve a high call rate for all selected SNPs, without time-consuming assay optimization processes, and at a relatively high multiplex level.
A key issue in high-throughput genotyping is to choose the appropriate technology for your goals and for the stage of your experiment. CD genomics offers researchers the flexibility to profile samples with thousands to millions of markers in high-throughput format, and deliver dense genome-wide coverage with the most up-to-date content available from the scientific community.
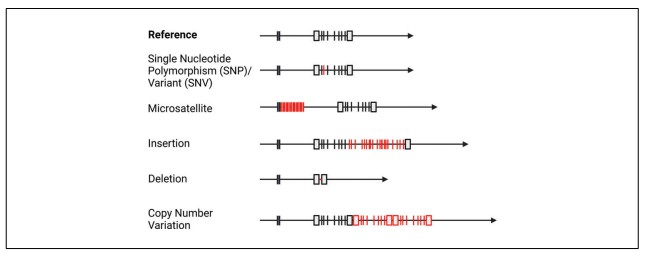 Figure 1. Types of genetic variants. (Kockum et al., 2023)
Figure 1. Types of genetic variants. (Kockum et al., 2023)
What are the Genotyping Method
Genotyping is a complex procedure that employs diverse techniques to dissect and characterize distinct genetic loci within an individual's genome. Here is an exhaustive synopsis of the constituent methodologies that make this process possible:
- Preparation of DNA:
Extraction of genomic DNA involves isolating chromosomal DNA from the nuclei of cells through the use of detergents and mechanical shearing. This is followed by the removal of proteins and cellular debris to acquire purified DNA. Extraction methods can be organic in nature, or they might utilize commercial kits harnessing filter columns or magnetic beads, processing biological samples like blood, saliva, or paraffin-embedded biopsy materials.
- Hybridization:
Genotyping methods such as PCR, microarrays, and NGS leverage hybridization, a process involving the pairing of complementary single-stranded DNA fragments to create double-stranded molecules, relying on base pair matching across these fragments, is fundamental.
- PCR-Based Techniques:
Polymerase Chain Reaction (PCR) facilitates the rapid amplification of specific DNA sequences, which aids in automated genotyping. Techniques like allele-specific PCR and PCR-RFLP are widely used in genotyping for the detection of specific alleles or distinguishing between alleles based on the patterns of restriction enzyme digestion. TaqMan PCR, known for its use of allele-specific probes with fluorescent labels, is prevalent for genotyping candidate SNPs.
- iPLEX:
The iPLEX methodology is based on Sequenom MassARRAY technology. It uses locus-specific primers and mass-modified dideoxynucleotide terminators for genotyping, which are detected with MALDI-TOF mass spectrometry.
- Pyrosequencing:
Pyrosequencing is employed for short DNA fragments. It involves the synthesis of novel DNA strands one nucleotide at a time, with light emission with each nucleotide incorporation, facilitating genotyping analysis.
- Microarray Techniques:
Microarrays, such as Affymetrix GeneChip and Illumina bead arrays, are a key part of broad-scale genotyping. They are based on the hybridization of DNA fragments to artificially synthesized DNA spots containing SNP sequences. Signal detection following this hybridization aids in genotype determination.
NGS platforms enable massively parallel sequencing of DNA fragments and enable high-throughput genotyping. This process incorporates a multitude of steps including sample preparation, cluster generation, sequencing-by-synthesis, and data analysis. It allows the identification and detection of novel polymorphisms and mutations. Within genotyping, the differences in DNA sequences across samples at the same locus are identified. Furthermore, in the case of the sample being RNA, NGS can quantify gene expression. The distinct edge of this methodology for genotyping is its capacity to identify novel polymorphisms/mutations, a capability that outshines other genotyping methods such as microarray.
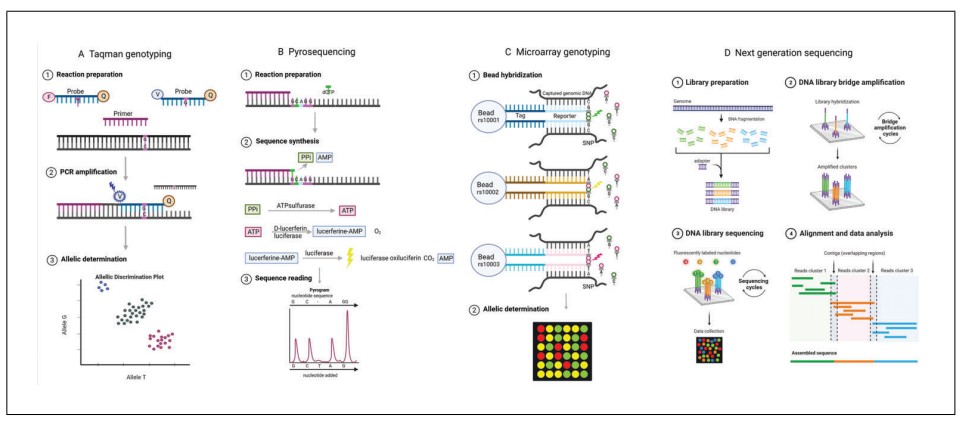 Figure 2. Selected genotyping methods. (Kockum et al., 2023)
Figure 2. Selected genotyping methods. (Kockum et al., 2023)
Our Genotyping services

We offer different genotyping technologies depending on the application and species, the purpose of the analysis, the number of SNPs per sample (from just a few up to five million) and the number of samples in the study. Our comprehensive SNP genotyping services include both whole genome wide assay for SNP screening and fine mapping assay for validation of SNPs based on 5 platforms—Affymetrix, Illumina, Sequenom MassARRAY, ABI TaqMan, and ABI 3730XL:
-
Whole Genome SNP Genotyping
-
Genotyping by Sequencing (GBS). We employ the latest Illumina sequencing platforms to obtain genetic variations information in a more efficient way. GBS, a subset of Reduced-Representation Genome Sequencing techniques, is employed to identify SNPs for genotyping investigations. GBS simplifies genome complexity by utilizing restriction enzymes to cleave DNA, coupled with DNA-barcoded adapters. Through judicious selection of restriction enzymes, GBS offers extensive SNP coverage in gene-rich regions of the genome in a cost-effective manner, enhancing tag numbers in the assay. The consistent presence of restriction sites across species ensures high genomic consistency within populations, rendering GBS particularly suitable for surveys.
SNP Microarray. Convergence of DNA hybridization, fluorescence microscopy, and solid surface DNA capture through both Affymetrix and Illumina Arrays. CD Genomics offers a wide range of genotyping services utilizing advanced variant detection and SNP genotyping technologies. Microarrays play a crucial role in exploring genetic variations such as SNPs and structural changes, supporting applications from initial discovery to routine screening. Affymetrix and Illumina genotyping microarrays are highly regarded for their consistent performance in various fields including precision medicine, clinical research, pharmacology, consumer screenings, and agriculture. These microarrays are customizable to specific species and SNP preferences, offering multiple genotyping strategies to meet diverse research needs
2b-RAD. A new cost-effective genome-wide genotyping method, 2b-RAD, uses type IIB restriction enzymes to generate fixed-size DNA fragments, targeting all restriction sites for uniform sequencing depth and high reproducibility. This method, combined with Illumina paired-end sequencing, offers advantages like low sequencing cost, resilience to DNA degradation, and flexibility in library design compared to traditional RAD and GBS techniques.
ddRAD-seq. ddRAD-seq, a favored tool among molecular ecologists, utilizes next-generation sequencing (NGS) platforms to create novel SNP markers. By exploiting the specificity of restriction endonuclease enzymes, it generates library fragments representing unique genomic regions shared among individuals of the same species. This enables efficient sequencing and comparison of these genomic regions across individuals, facilitating rapid and effective development of numerous genetic markers.
PCR-LDR SNP Genotyping. High-throughput molecular marker technologies like SNP chips and Genotyping-By-Sequencing (GBS) have broad applications in genetics research, including SNP discovery, crop breeding, and genome-wide association studies. However, their cost and complexity limit their use in crop breeding. Instead, PCR-based molecular markers targeting functional polymorphic loci are commonly employed. These markers utilize Taq DNA ligase to detect variations in genotypes, enabling precise SNP site detection.
RAD-seq. RAD-seq, an innovative next-generation sequencing (NGS) technique, is designed to target specific genomic cleavage sites using restriction endonucleases, resulting in the generation of predefined DNA fragments suitable for high-throughput sequencing. Its adaptability renders it well-suited for comprehensive investigations, providing valuable contributions to the fields of SNP identification, genetic mapping, functional genomics, population genetics, and beyond, thereby offering significant implications for both research endeavors and industrial applications.
-
-
SNP Fine Mapping
-
MassARRAY SNP Genotyping. A non-fluorescent detection platform utilizing mass spectrometry to accurately and sensitively measure PCR-derived amplicons.
SNaPshot. A primer extension-based method for genotyping known SNP positions through the automated DNA analyzer.
TaqMan SNP Genotyping. A commonly used SNP genotyping method developed by Life Technologies.

-
-
CNV Genotyping
-
CGH Microarray. Microarray CGH enables comprehensive genome-wide screening at high resolution, facilitating the detection of previously undetected copy number imbalances such as deletions and duplications. CD Genomics offers CGH microarray services for various species, encompassing human, mouse, rat, and chicken, with the option for custom CGH arrays tailored to other species.
-
-
DNA Fragment Service
-
Microsatellite Genotyping. CD Genomics specializes in microsatellite marker selection and design across diverse species. Alongside microsatellite genotyping, we offer efficient genotyping by sequencing (GBS) to optimize time and resources. Leveraging PCR and next-generation sequencing, genotyping ensures accurate, cost-effective allele differentiation.
Microsatellite Instability Analysis. Microsatellite instability (MSI) arises from errors in DNA replication, where repeating bases (microsatellites) are inserted or deleted due to mismatch repair system (MMR) failure. CD Genomics employs the Promega kit, utilizing fluorescent multiplex PCR-Capillary Electrophoresis, to detect MSI status.
Microsatellite Development. CD Genomics now offers a next-generation sequencing-based method for developing microsatellite markers tailored to your species of interest, contributing to scientific advancement. Microsatellites, or SSRs, are repetitive DNA sequences prevalent in eukaryotic genomes, characterized by high polymorphism and suitability for various genetic studies. NGS revolutionizes microsatellite marker development, offering high-throughput, cost-effective analysis even for non-model organisms. Utilizing Illumina technology, millions of short fragment reads are generated and screened bioinformatically to identify polymorphic loci.
Hi-SSRseq. Microsatellites, or SSRs, are essential genetic markers in population genetics. However, traditional genotyping methods based solely on fragment length may overlook SNPs and indels, leading to inaccurate results. CD Genomics offers Hi-SSRseq, combining SSR amplification with high-throughput sequencing for accurate genotyping. This method provides nucleotide sequence data for SSR and flanking regions, enabling precise assessment of genetic variability and reducing genotyping time and cost.
-
Sample requirements
| Service | Sample Type | Recommended Quantity | Minimum Quantity | Minimum Concentration |
| GBS/ddRAD | Genomic DNA | ≥ 300 ng | 100 ng | 10 ng/µL |
| 2b-RAD | Genomic DNA | ≥ 200 ng | 50 ng | 5 ng/µL |
| SNP Microarray | Genomic DNA | ≥ 300 ng | 100 ng | 8 ng/µL |
| SSR Genotyping | Genomic DNA | ≥ 500 ng | 200 ng | 10 ng/µL |
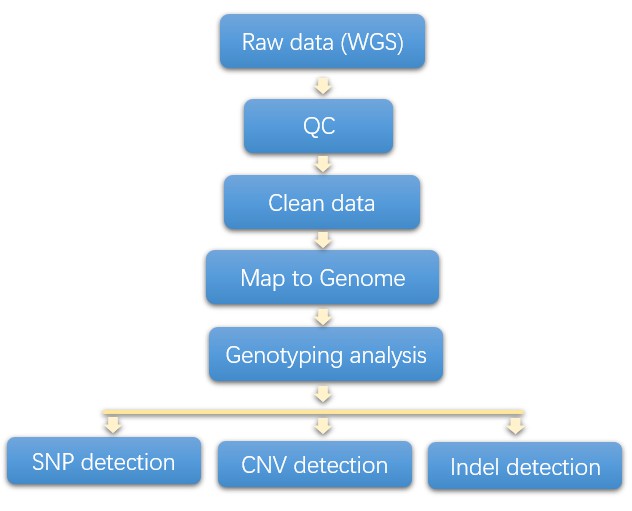
Analysis Pipeline
The fundamental input data can comprise diverse types of microarray chip data and next-generation sequencing data such as whole genome sequencing or whole exome sequencing data. Ordinarily, genotyping results from microarray chip data are furnished by the chip manufacturer. The subsequent pipeline delineates the systematic approach to analyzing genotyping by utilizing whole genome sequencing data.
Advantages of Our Genotyping Services
- Rigorous adherence to stringent standards ensures the delivery of comprehensive genotyping solutions.
- Expertise across various facets of genotyping, including assay design, sample processing, and data analysis, guaranteeing accurate and insightful results.
- Swift turnaround time coupled with unwavering accuracy and reliability make our genotyping services both efficient and dependable.
- Versatility to handle diverse sample types, spanning human, animal, plant, and microbial genomes, underscores our capability to address a broad spectrum of research needs.
- Commitment to advancing genomics research by offering access to cutting-edge genotyping technologies and methodologies.
- Tailored services tailored to suit the unique requirements of each project, ensuring a personalized and effective approach to genotyping analysis.
- Consultative approach to identify optimal and cost-effective solutions, empowering researchers to achieve their specific research objectives with maximum efficiency.
Our service package includes:
- Scalable platform. CD Genomics is able to perform an effective analysis on small to large samples due to its variety of SNP genotyping platforms.
- Quality assurance. Through our strict QC process, we only provide data of high resolution and high quality for the utmost reliability.
- Experiment design. Selection of SNP fit for research purpose, and then a selection of platform based on the number of samples and design possibility.
- Genotyping. Validation of the existence and frequency of known SNPs and discovery or novel SNPs.
- Data. Provided in Excel format. For SNP discovery, raw data, annotation, LD results are also provided.
Results Display
The application of bioinformatics approaches to the analysis of SNP genotyping and Copy Number Variations (CNVs) allows for the examination of results from millions of markers and probes, the detection of outlier samples, and the prediction of functional consequences of genetic variations. Presented below are graphical representations of the outputs generated using assorted genotyping techniques.
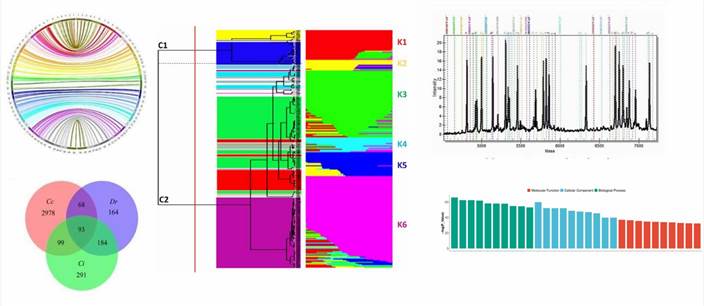
We aim to provide the highest level of service, confidentiality and customer support. Our extensive expertise can help expedite your SNP and mutation analysis assays with speed, quality, and reliability. With our services you can either take a deep look into the genome or search for unknown genotype-phenotype connections or genotype already known variations.
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Cells | 2021MLST-Based Analysis and Antimicrobial Resistance of Staphylococcus epidermidis from Cases of Sheep Mastitis in Greece
Biology | 2021Increased Production of Pathogenic, Airborne Fungal Spores upon Exposure of a Soil Mycobiota to Chlorinated Aromatic Hydrocarbon Pollutants
Microbiology Spectrum | 2023Pinpointing Rcs3 for frogeye leaf spot resistance and tracing its origin in soybean breeding
Molecular Breeding | 2023

 Sample Submission Guidelines
Sample Submission Guidelines
