ctDNA Sequencing Services
Introduction of ctDNA Sequencing
Circulating tumor DNA (ctDNA) is a type of extracellular DNA present in body fluids such as plasma, serum, cerebrospinal fluid, etc. It primarily originates from necrotic or apoptotic tumor cells, extracellular vesicles secreted by tumor cells, and circulating tumor cells. The typical size of ctDNA is in the range of 160-180 base pairs. ctDNA is a subset of cell-free DNA (cfDNA) and constitutes a relatively low proportion (between 0.1% and 1%), making its detection challenging. The maturation of next-generation sequencing (NGS) technology has significantly enhanced the sensitivity and accuracy of ctDNA detection.
Circulating cell-free DNA (cfDNA) primarily consists of lineage DNA from normal cells, where circulating tumor DNA (ctDNA) in cancer patients is relatively scarce and greatly variable. Consequently, conventional DNA analysis methods such as Sanger sequencing and pyrophosphate sequencing lack the sensitivity required to detect somatic mutations in plasma ctDNA of cancer patients. Currently, there are three main analysis methods for ctDNA: quantitative PCR techniques represented by ARMS, digital PCR techniques represented by ddPCR, and high-throughput detection techniques based on Next-Generation Sequencing (NGS). In recent years, nucleic acid mass spectrometry and EFIRM technologies have also emerged.
To enhance detection throughput and efficiency, sequencing technologies centered around NGS are widely used for downstream analysis of ctDNA, ranging from whole-genome or whole-exome sequencing to targeted sequencing of a limited genome. NGS enables high-throughput parallel deep sequencing of multiple gene nucleic acid segments, facilitating simultaneous detection of multiple genes, various forms (such as mutations, copy number changes, and gene fusions), and unknown mutations. The involvement of multiple genes in prognosis and treatment efficacy assessment could have a significant impact on prognostic outcomes.
Characteristics of Circulating Tumor DNA Sequencing
Convenient Sampling: Non-invasive, real-time, easily accessible multiple times ctDNA analysis is achieved by extracting blood, eliminating the risks and discomfort associated with invasive methods such as biopsies and surgeries for collecting tumor tissue. In the early stages of malignant tumors, alterations in ctDNA content and genes can be detected.
Mature Detection Technology: High sensitivity, high accuracy, low false-positive rate Utilizing NGS for ctDNA detection ensures high sensitivity and accuracy, detecting mutations as low as 0.1%. Compared to other types of tumor markers, ctDNA exhibits higher sensitivity, making it an ideal tumor biomarker.
Comprehensive Detection: Broad Application Range Nearly all tumor cells release DNA fragments into the bloodstream. Consequently, ctDNA reflects the overall status of tumors in the body. Heterogeneity significantly impacts gene testing in tumor tissues, but ctDNA detection mitigates the randomness and locality associated with tumor tissue heterogeneity. This ensures more comprehensive and reliable results for tumor mutations.
Advantages of ctDNA Sequencing
- Comprehensive Service: We offer an all-inclusive service, covering the entire process from ctDNA extraction, library preparation, sequencing, to data analysis.
- Ultra-Low ctDNA Input: Our technology allows for sequencing with starting ctDNA amounts as low as 10 ng.
- Stringent Quality Control: A rigorous quality control process is implemented at every stage, ensuring high accuracy in the results provided to our clients.
- Specialized Bioinformatics Analysis: Our proficient bioinformatics team is equipped to meet the personalized data analysis requirements of our clients.
Application of ctDNA Sequencing
- Detection of Early-Stage Cancer: By examining circulating tumor DNA (ctDNA) in the bloodstream, it becomes feasible to identify tumor-specific mutations in the nascent stages, thereby improving the prospects of early diagnosis.
- Cancer Monitoring: The dynamic monitoring of ctDNA levels in patients offers insights into variations in tumor burden, providing a means to evaluate treatment efficacy and disease progression.
- Recurrence Prediction: The resurgence or elevation of ctDNA levels in postoperative or post-treatment follow-ups can potentially forecast cancer recurrence.
- Personalized Therapy: Through the analysis of ctDNA sequencing outcomes, clinicians can design tailored treatment regimens, opting for targeted therapies or modifying treatment strategies accordingly.
Our Provided ctDNA Sequencing Service
Whole-Genome Resequencing for ctDNA
We provide comprehensive whole-genome resequencing for ctDNA, enabling the analysis of various types of mutations carried by ctDNA, such as single nucleotide polymorphisms (SNPs), insertions and deletions (indels), and large structural variations.
Exome Sequencing for ctDNA
Similar to conventional genomic DNA, a sufficient quantity of ctDNA can undergo exome sequencing to detect variations in coding regions.
Methylation Sequencing for ctDNA
Methylation sequencing for ctDNA is a focal point in research. This includes the detection of methylation sites for individual genes and comprehensive methylation site detection across the entire genome.
Targeted Region Sequencing for ctDNA
Our targeted region panel for ctDNA allows for the detection and analysis of specific target sites. In comparison to whole-genome sequencing, this method provides a more cost-effective solution.
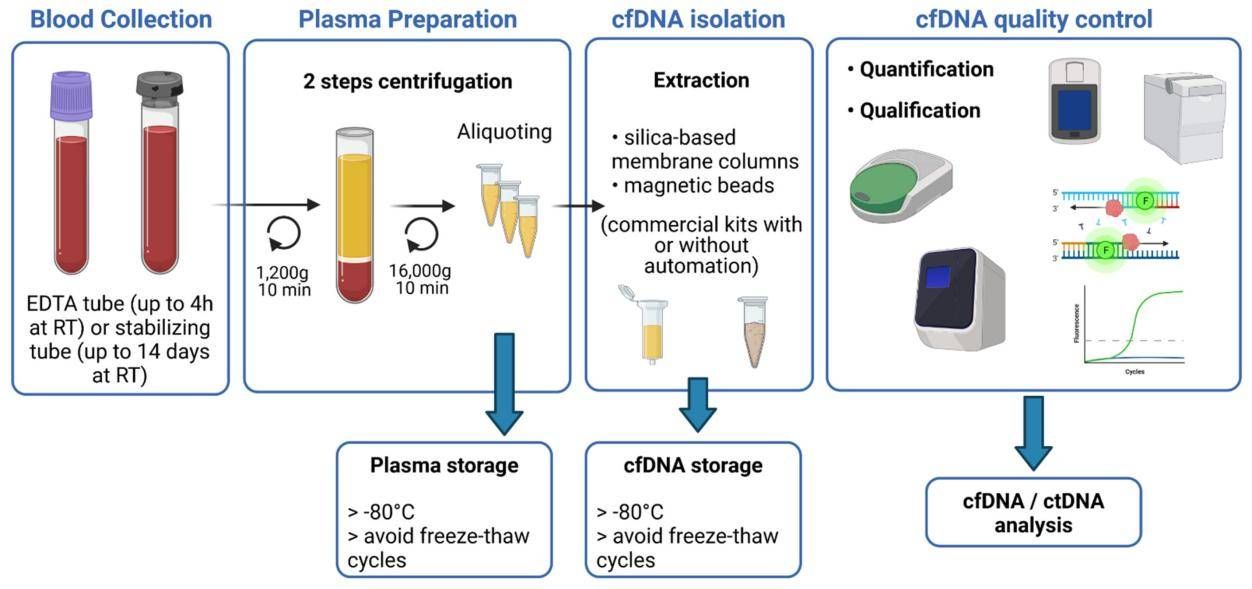 Figure 1. Schematic overview of the main steps for blood sample processing and cfDNA extraction. (Bohers et al,. 2021)
Figure 1. Schematic overview of the main steps for blood sample processing and cfDNA extraction. (Bohers et al,. 2021)
ctDNA Sequencing Workflow
CD Genomics provides fast and accurate ctDNA sequencing services and bioinformatics analysis.

Service Specifications
Sample Requirements
Plasma is recommended over serum to minimize interference from lymphocyte-derived cfDNA.
2. If low-temperature centrifugation is not possible, use STRECK non-invasive tubes with a proprietary stabilizer for whole blood preservation, allowing room temperature centrifugation. 3. To eliminate residual cells, plasma should undergo a secondary centrifugation: first at 4℃ at 1600g for 10 minutes, followed by a second centrifugation at 4℃ at 16000g for 10 minutes.
|
|
Click |
Sequencing Strategy
|
Bioinformatics Analysis
|
Analysis Pipeline
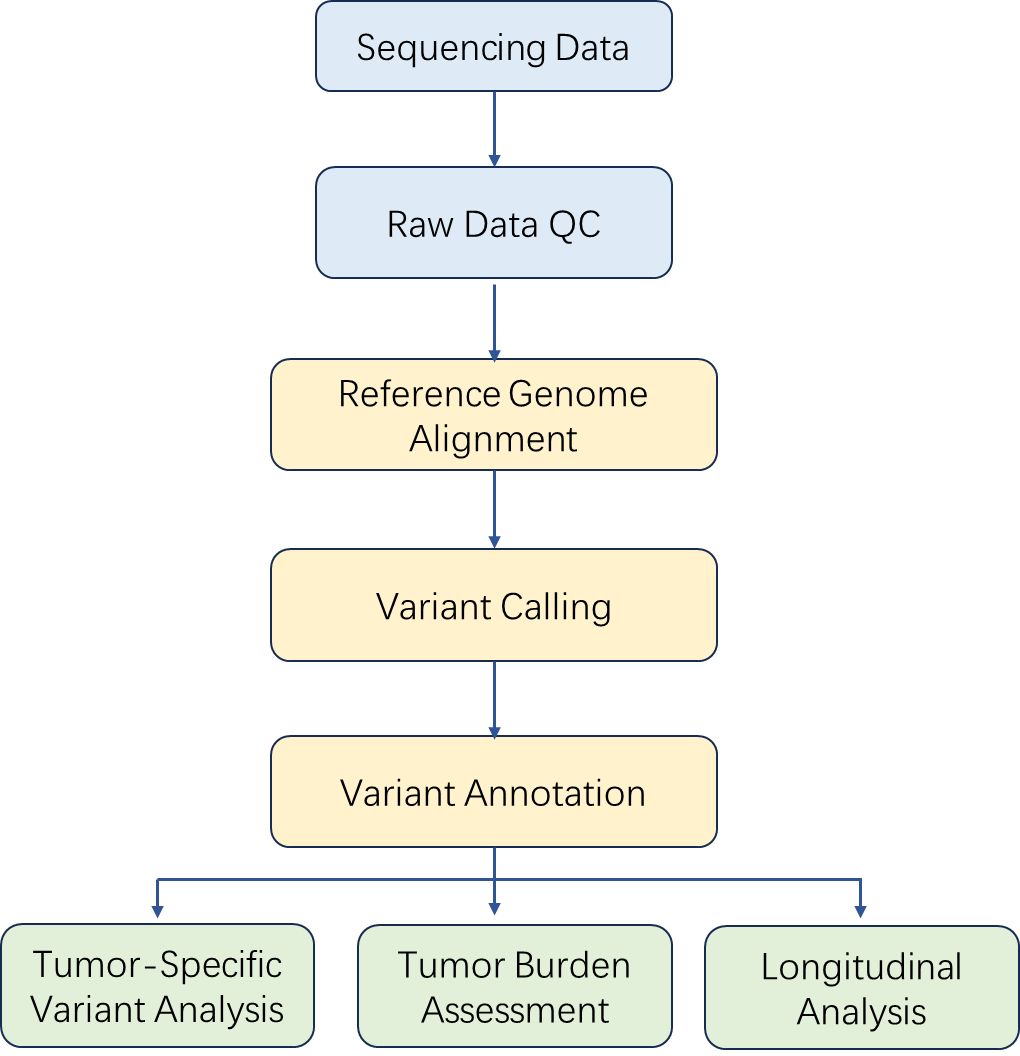
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in ctDNA Sequencing for your writing (customization)
References
- Michail Ignatiadis, et al. Liquid biopsy enters the clinic-implementation issues and future challenges. Nat Rev Clin Oncol. 2021 M
- Ming Chen, Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019 Aug 1;13(1):34.
- Carolin M Sauer, Longitudinal monitoring of disease burden and response using ctDNA from dried blood spots in xenograft models. EMBO Mol Med. 2022
- Etienne Giroux Leprieur, Sequential ctDNA whole-exome sequencing in advanced lung adenocarcinoma with initial durable tumor response on immune checkpoint inhibitor and late progression. J Immunother Cancer. 2020
- Anjui Wu, Genome-wide plasma DNA methylation features of metastatic prostate cancer. J Clin Invest. 2020
- Jianxia Chen, et al. Design of a Targeted Sequencing Assay to Detect Rare Mutations in Circulating Tumor DNA. Genet Test Mol Biomarkers. 2019
- Bohers E, Viailly P J, Jardin F. cfDNA sequencing: technological approaches and bioinformatic issues. Pharmaceuticals, 2021, 14(6): 596.
Demo Results
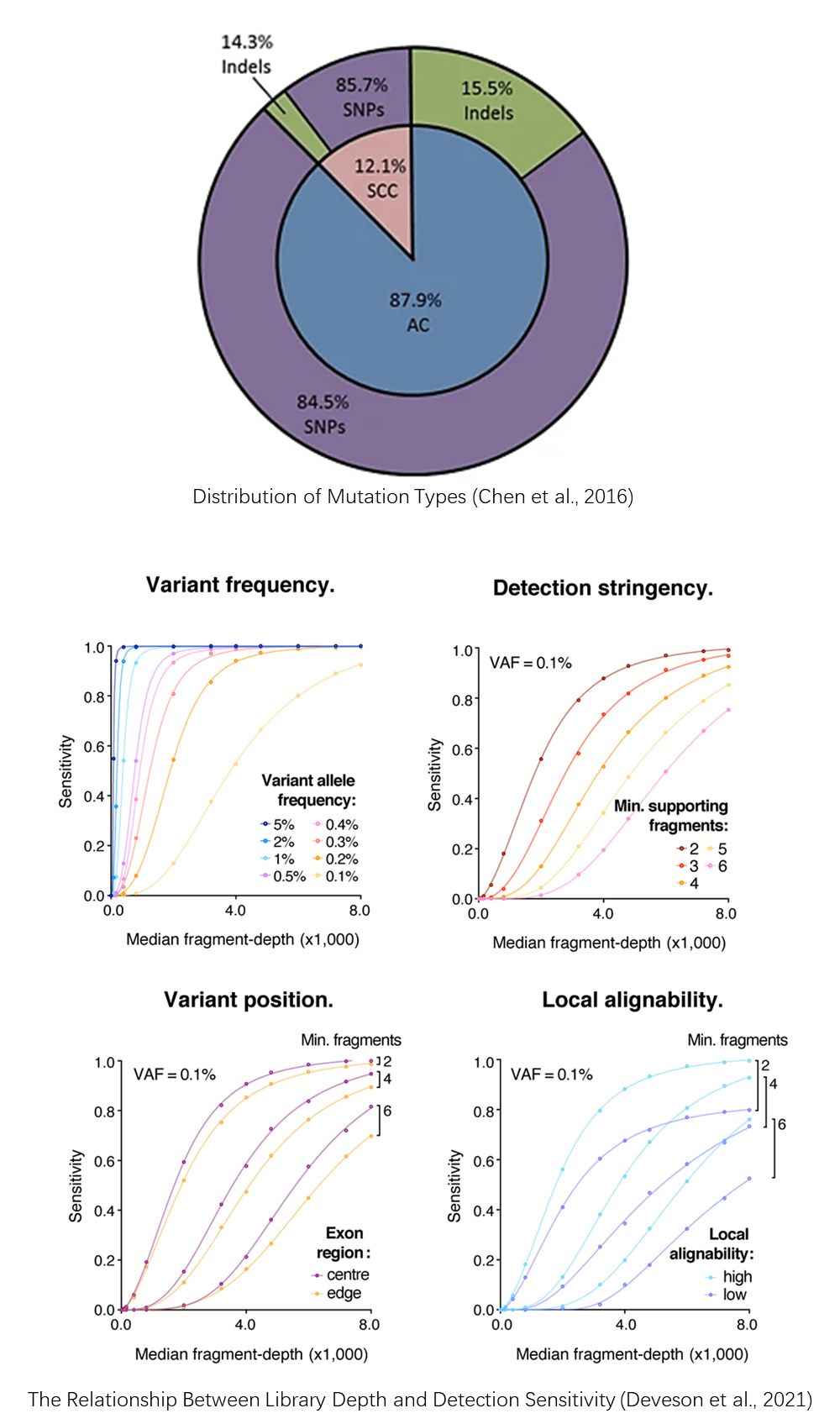
References
- Chen, KZ., Lou, F., Yang, F. et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci Rep, 2016, 6, 31985.
- Deveson I W, Gong B, Lai K, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nature biotechnology, 2021, 39(9): 1115-1128.
ctDNA Seq FAQs
1. What are the advantages and disadvantages of ctDNA compared to histological testing?
- Advantages: In comparison to histological testing, ctDNA analysis offers non-invasive or minimally invasive methods, facilitates repeated sampling, has shorter collection, processing, and reporting turnaround times. It can overcome spatial tumor heterogeneity and provide real-time, relatively comprehensive insights into the molecular characteristics of a patient's tumor. Additionally, plasma ctDNA can also increase the detection rate of driver gene mutations.
- Disadvantages: In contrast to tissue samples, peripheral blood ctDNA has lower content, which can lead to false-negative clinical test results. The presence of germline variations or clonal hematopoiesis mutations might also cause false positives if white blood cells are not used as a control. Moreover, variations exist in the amounts of ctDNA released into the bloodstream by different cancer patients or the same patient at different time points, posing challenges for clinical testing and result interpretation.
2. Why does ctDNA testing require higher sensitivity?
The number of tumor cells in the body of a cancer patient is significantly lower than that of normal cells, and cfDNA levels in plasma are generally low. ctDNA comprises only 0.1% to 5% of cfDNA, with substantial differences in ctDNA levels in plasma among different cancer types and disease stages. Therefore, compared to tissue testing, ctDNA detection necessitates higher sensitivity and specificity.
Current technologies used for liquid biopsy of ctDNA predominantly include ARMS-PCR, digital PCR (ddPCR), and next-generation sequencing (NGS).
3. What is non-targeted sequencing in ctDNA analysis?
Non-Targeted Sequencing in ctDNA analysis refers to the approach of examining ctDNA without a predetermined focus on specific alterations, a methodology that encompasses Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS). WGS, in particular, has been utilized for ctDNA analysis due to its capacity to unveil tumor-specific variations without prior knowledge of potential mutations. This allows for the identification of gene alterations associated with resistance to treatment and the pinpointing of newly actionable targets. Nonetheless, WGS comes with drawbacks such as lower sensitivity rates of 5-10%, elevated expenses, prolonged detection periods, and increased complexity in data analysis, rendering its integration into clinical practice a formidable task.
4. How is targeted sequencing performed in ctDNA analysis?
Targeted sequencing can enrich target fragments through PCR or hybrid capture. PCR-based targeted sequencing designs dozens to hundreds of PCR primers targeting specific genes for enrichment. Hybrid capture-based targeted sequencing designs probes targeting specific genes and enriches them through hybrid capture.
5. What strategies are used to improve sensitivity and specificity in ctDNA analysis?
Due to the complexity of next-generation sequencing (NGS) workflows, errors introduced during library construction, target fragment enrichment, and sequencing inevitably lead to background noise. This background noise can obscure low-frequency mutations in ctDNA samples, leading to false-negative or false-positive results, limiting the sensitivity and specificity of ctDNA detection. Therefore, most NGS-based targeted sequencing technologies aim to reduce background noise and improve sensitivity and specificity, often using molecular barcoding strategies. Initially, single-stranded DNA barcodes were used, but now, most strategies use double-stranded DNA barcodes. While double-stranded barcodes offer better error suppression, they have relatively lower efficiency, making them less optimal for limited cfDNA samples in clinical settings.
ctDNA Seq Case Studies
Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma
Journal: Clinical cancer research
Impact factor: 10.86
Published: 1 December 2019
Background
Gastroesophageal adenocarcinoma (GEA) is a significant global health problem with limited survival rates despite optimal treatments. Although tissue-based next-generation sequencing (NGS) has identified key genomic alterations (GAs), intrapatient heterogeneity complicates targeted therapy. Recent studies suggest that ctDNA-NGS, offering a noninvasive method, may better guide targeted therapy selection by providing a comprehensive view of GAs and overcoming some limitations of tissue-NGS. This study analyzes a large cohort of GEA patients using ctDNA-NGS to evaluate its detection limits, correlation with clinical outcomes, and potential to predict therapy response and resistance.
Methods
Sample Preparation:
1,630 patients
GEA samples
Sequencing:
CtDNA-NGS
Tumor location
Genetic landscape
ctDNA as a biomarker
Heterogeneity between disease sites
Results
Plasma cfDNA assays rely on the release of tumor DNA (ctDNA) into the bloodstream, mixing with normal cfDNA. The maximal tumor somatic variant allelic frequency (maxVAF) reflects the largest ctDNA clone and helps estimate overall ctDNA quantity and subclonality. In untreated stage IV patients, higher maxVAF correlates with greater disease burden and specific metastatic sites, indicating that both disease location and extent significantly affect ctDNA shedding and detection sensitivity.
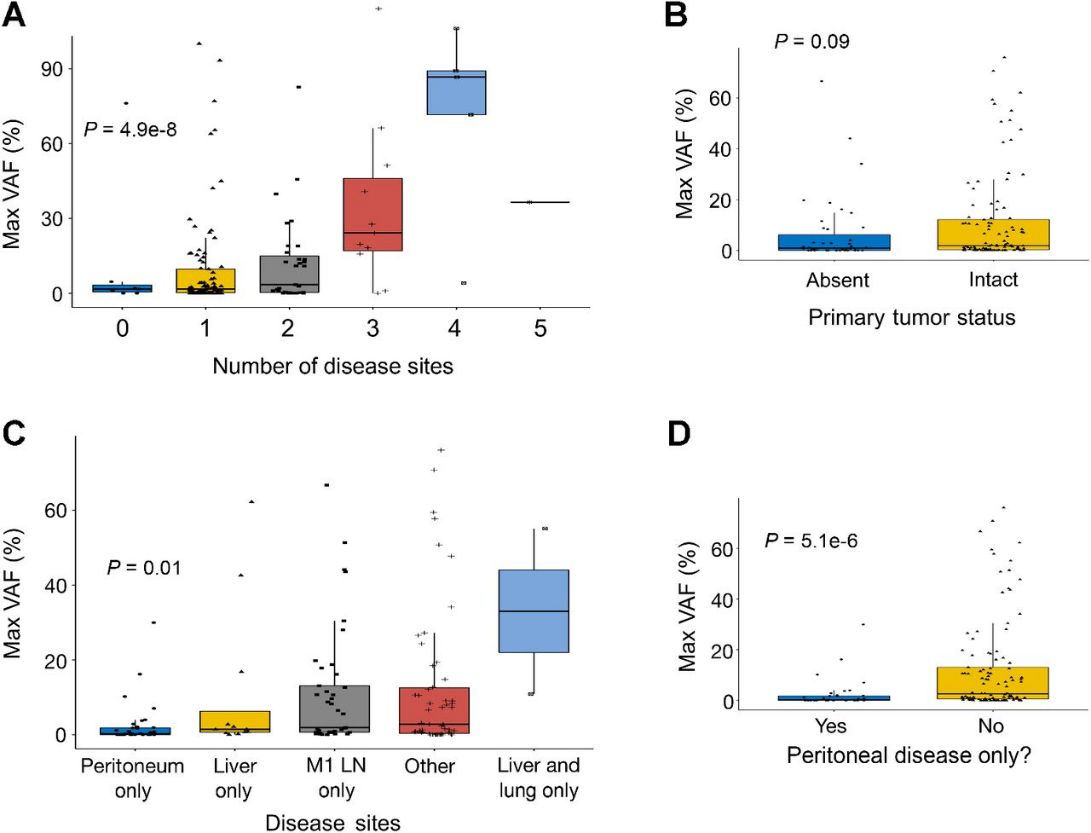 Fig 1. ctDNA detection and number of detected alterations is dictated by specific disease sites and burden of disease.
Fig 1. ctDNA detection and number of detected alterations is dictated by specific disease sites and burden of disease.
Clinical ctDNA-NGS is used to identify actionable genomic alterations (GAs) and can potentially serve as a prognostic biomarker for both early- and late-stage disease. In locally advanced patients, detectable ctDNA was associated with shorter disease-free survival, and post-surgery ctDNA levels predicted recurrence. In advanced stage IV patients, higher ctDNA levels (maxVAF) correlated with worse prognosis. Serial ctDNA-NGS analysis indicated that a significant decline in maxVAF during first-line therapy predicted better survival. Among patients treated with immune checkpoint inhibitors, lower maxVAF was linked to longer overall survival.
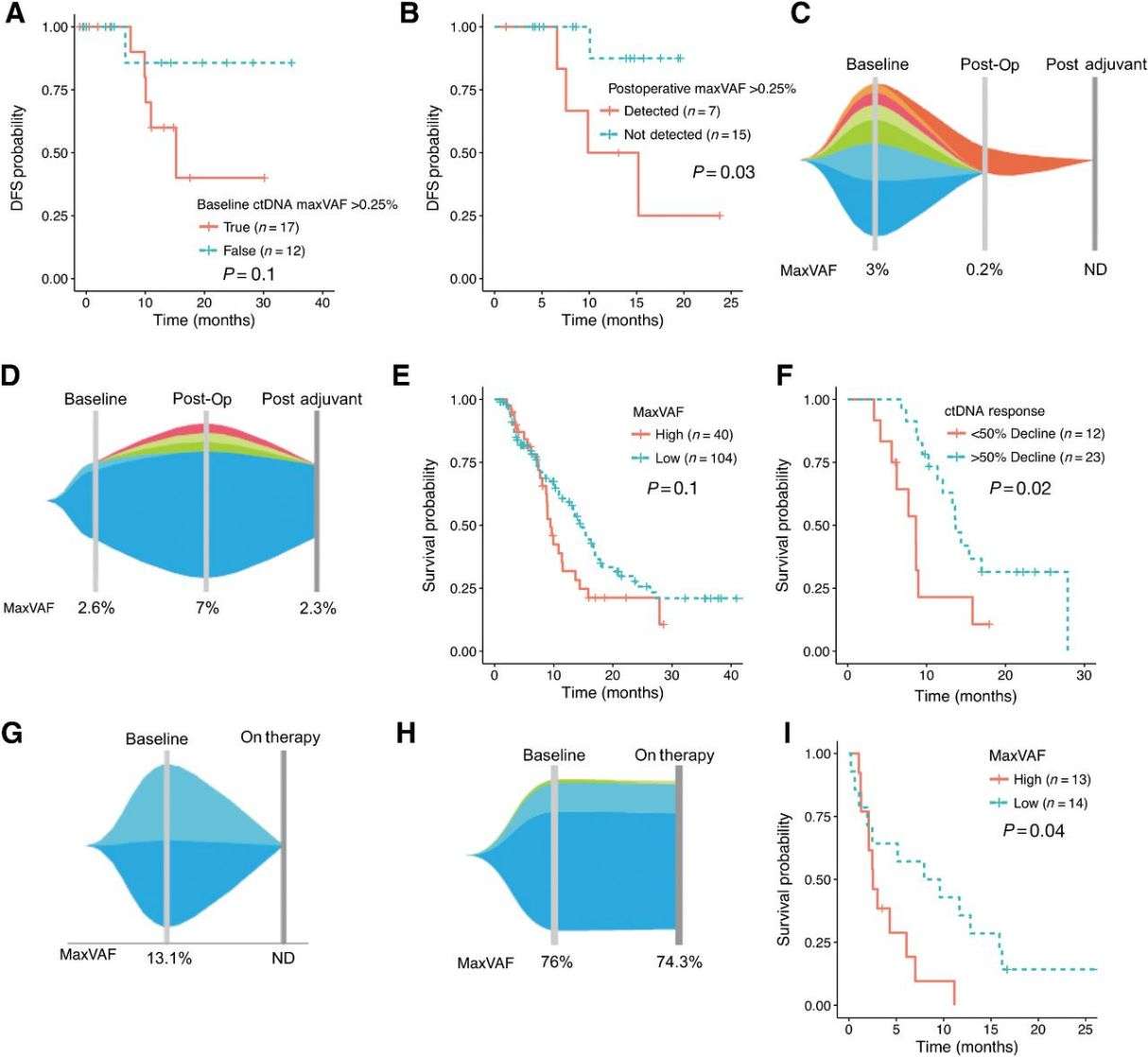 Fig 2. Prognostic implications of maxVAF and serial changes in the perioperative and newly diagnosed metastatic settings.
Fig 2. Prognostic implications of maxVAF and serial changes in the perioperative and newly diagnosed metastatic settings.
At initial diagnosis, spatial heterogeneity of HER2 and other genomic alterations (Gas) in GEA patients is significant. In a study of 34 untreated stage IV GEA patients, ctDNA-NGS and tissue-NGS from primary and metastatic sites showed that only 26% of Gas were universally concordant across samples, highlighting the complementary value of both methods. CtDNA-NGS also detected temporal heterogeneity and acquired resistance mutations post-therapy, which can guide optimal subsequent treatments. This underscores the importance of combining ctDNA-NGS with tissue-NGS for a comprehensive assessment of tumor heterogeneity and resistance mechanisms.
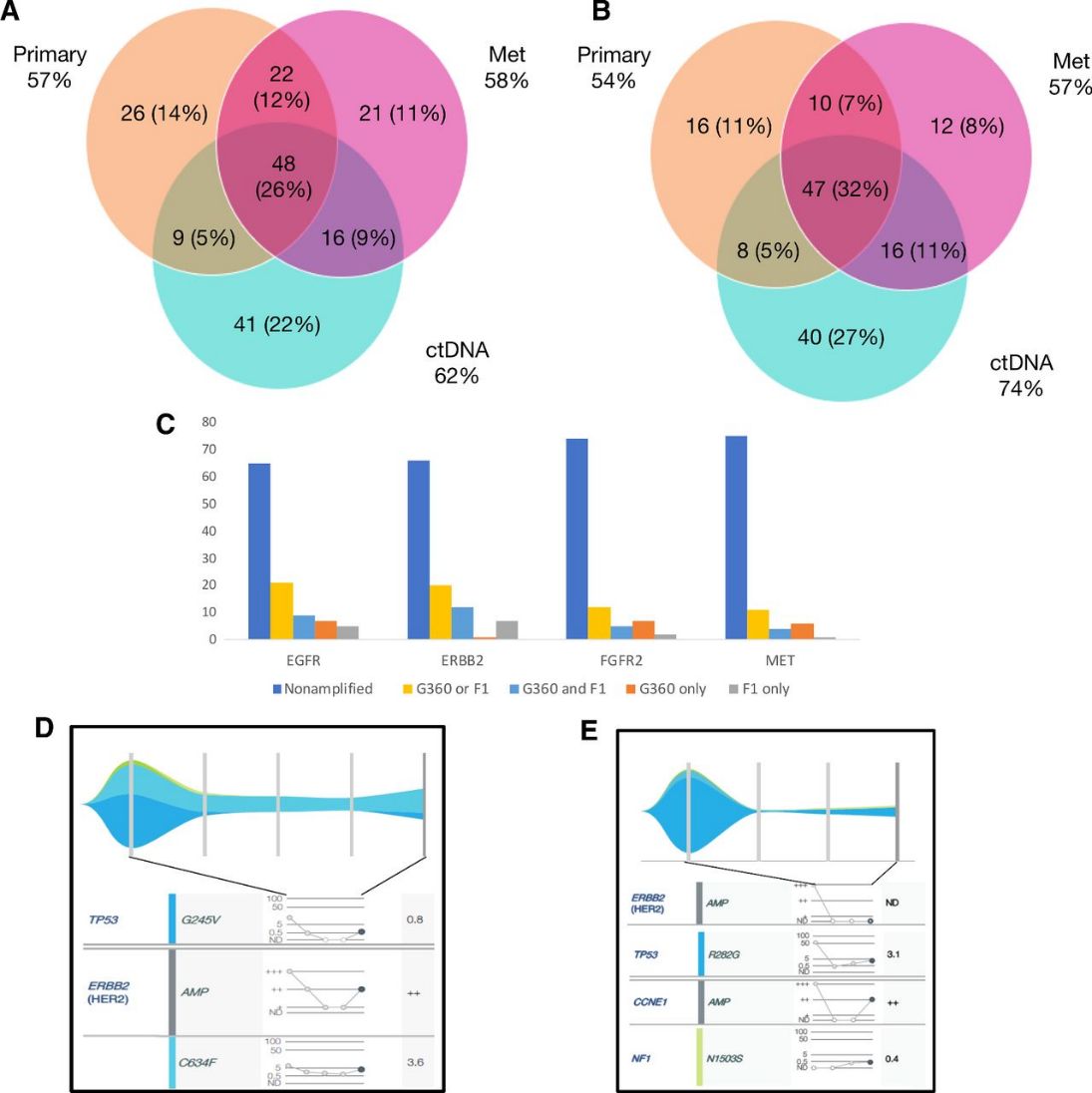 Fig 3. Intrapatient spatial and temporal heterogeneity by multisite tissue-NGS and ctDNA-NGS.
Fig 3. Intrapatient spatial and temporal heterogeneity by multisite tissue-NGS and ctDNA-NGS.
ctDNA-NGS can identify prognostic and predictive genomic alterations (GAs) in GEA patients. Mutations in PIK3CA and BRAF are associated with poor survival, while HER2 and EGFR amplifications, when identified by both ctDNA-NGS and tissue-NGS, predict better outcomes with targeted therapies. Combining ctDNA-NGS with tissue-NGS enhances detection of GAs, providing a more comprehensive assessment for guiding treatment strategies.
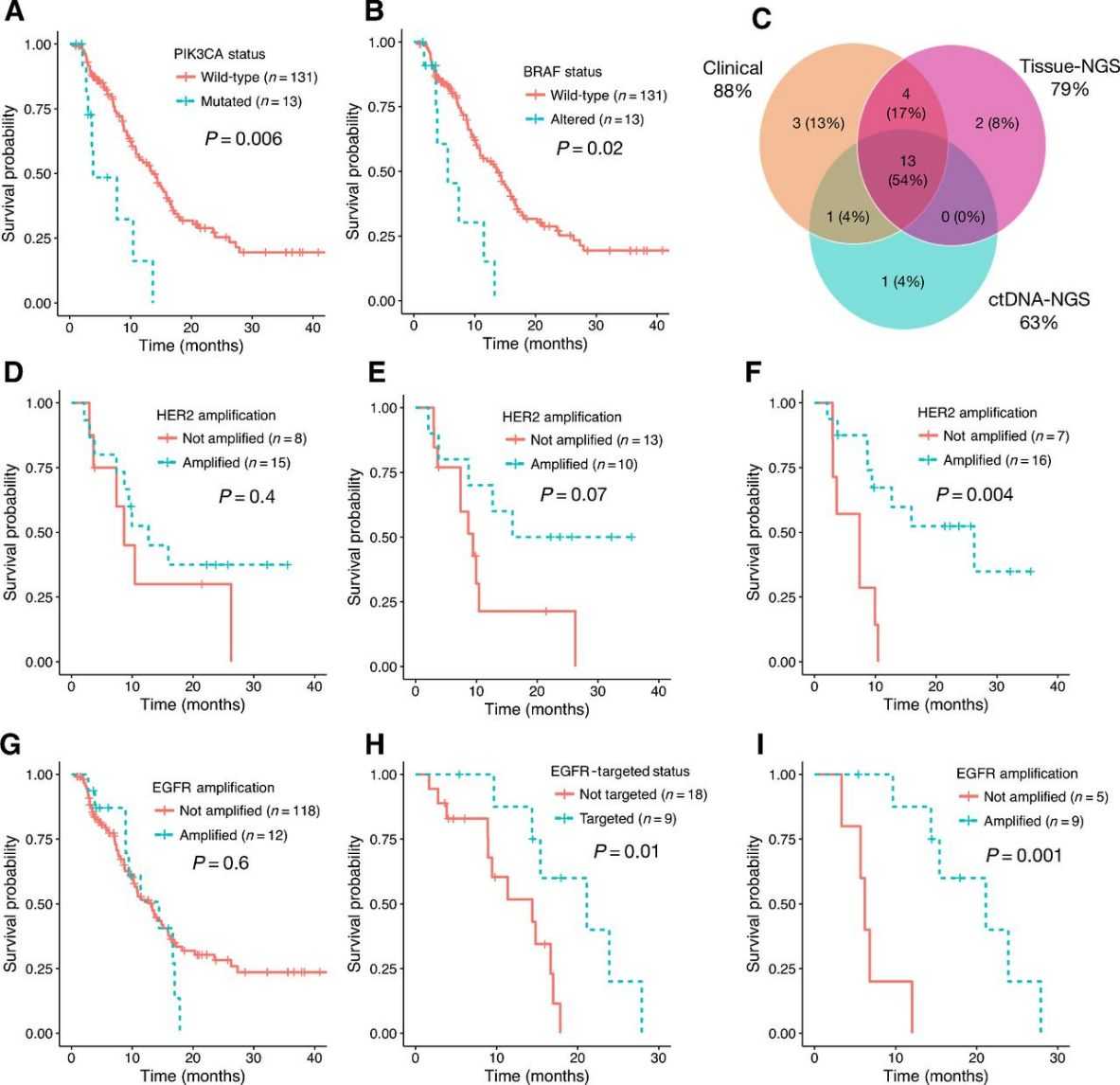 Fig 4. Survival analysis of untreated stage IV GEA patients by specific GA.
Fig 4. Survival analysis of untreated stage IV GEA patients by specific GA.
Conclusion
This study, the largest of its kind, used ctDNA-NGS to analyze 1,630 GEA patients. It established detection limits, confirmed the prognostic value of residual ctDNA post-surgery, and mapped the GA landscape, showing ctDNA-NGS accurately identifies actionable GAs and resistance mechanisms. Combining ctDNA-NGS with tissue-NGS improves detection, suggesting ctDNA-NGS can enhance monitoring and treatment of GEA, though further validation is needed.
Reference
- Maron S B, Chase L M, Lomnicki S, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clinical cancer research, 2019, 25(23): 7098-7112.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Distinct functions of wild-type and R273H mutant Δ133p53α differentially regulate glioblastoma aggressiveness and therapy-induced senescence
Journal: Cell Death & Disease
Year: 2024
High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing
Journal: International Journal of Molecular Sciences
Year: 2020
Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI
Journal: New phytologist
Year: 2017
Generation of a highly attenuated strain of Pseudomonas aeruginosa for commercial production of alginate
Journal: Microbial Biotechnology
Year: 2019
Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics
Journal: Viruses
Year: 2024
Genome Analysis and Replication Studies of the African Green Monkey Simian Foamy Virus Serotype 3 Strain FV2014
Journal: Viruses
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
