What Is Multi-Omics Analysis?
Multi-omics analysis is a systems biology approach that integrates multiple molecular layers—genomics, transcriptomics, epigenomics, proteomics, and metabolomics—to generate a comprehensive view of biological mechanisms. Unlike single-omics studies that focus on one molecular domain, multi-omics captures cross-talk between genes, transcripts, proteins, and metabolites, enabling more accurate modeling of biological function, disease progression, and therapeutic response.
Whether you're uncovering regulatory circuits in cancer, optimizing crop traits, or profiling host–microbe interactions, multi-omics data analysis delivers a deeper, more reproducible understanding of the system.
Key Advantages of Multi-Omics Approaches:
- Decodes complex interactions beyond the limits of single-omics
- Cross-validates results across biological layers for improved accuracy
- Enables discovery of multi-modal biomarkers and pathway signatures
- Offers high sensitivity to detect low-abundance but critical targets
- Supports both hypothesis-driven and discovery-based research
Why Multi-Omics Is Transforming Modern Bioscience
Integrating diverse omics data has become essential in both academic and industrial settings. From biomedical discovery to agricultural genomics and environmental science, multi-omics integration enables more precise research conclusions, accelerating innovation and reducing development risk.
Biomedical Research & Drug Development
- Identify disease-specific drivers across genomic, transcriptomic, and epigenomic layers
- Predict therapeutic targets and resistance mechanisms
- Track treatment response and immune modulation using longitudinal omics
Plant, Microbiome & Environmental Studies
- Improve stress tolerance and yield traits through omics-guided breeding
- Uncover soil–plant–microbiome interactions using amplicon + metabolomics integration
Monitor environmental pollutants through multi-level biomolecular profiling
Multi-Omics Integration Strategies
Transcriptomics + Metabolomics
This dual-layer strategy links gene expression to downstream metabolic changes, offering both causative and phenotypic perspectives. Ideal for:
- Growth & development studies
- Immune response profiling
- Stress resistance research
- Biomarker discovery in chronic disease
Transcriptomics + Proteomics
Explore the full cascade of gene regulation by analyzing mRNA and protein expression together. This approach captures both transcriptional activity and post-transcriptional modulation, uncovering hidden regulatory events.
- Reveal translation bottlenecks and compensatory pathways
- Compare gene-to-protein correlations under stress or disease
- Improve biomarker validation with protein-level evidence
Epigenomics + Transcriptomics + Genomics
This tri-omics approach offers a panoramic view of gene regulation—connecting sequence variants, epigenetic modifications (like DNA methylation and histone marks), and downstream gene expression.
- Map regulatory regions controlling gene activation
- Characterize cancer subtypes and developmental stages
- Identify disease-linked variants in non-coding regions
Popular Tools:
WGBS, RRBS, ChIP-seq, RNA-seq, and whole-genome sequencing (WGS)
Amplicon Sequencing + Metabolomics
Specially designed for microbiome studies, this combination uncovers how microbial populations influence host or environmental metabolism.
- Study gut microbiome modulation of host immune pathways
- Track soil microbial communities driving nitrogen cycling
- Identify microbial metabolites correlated with disease states
Example Use Case:
Linking 16S rRNA sequencing of gut flora with metabolomic shifts in short-chain fatty acid production.
Each project is supported by a dedicated bioinformatics team that will help you:
- Select optimal omics combinations
- Design a study that supports both discovery and validation
- Integrate public data (e.g., TCGA, GEO) to enhance interpretability
Customized Bioinformatics & Data Analysis
From Raw Multi-Omics Data to Biological Insight
At CD Genomics, we recognize that sequencing is only the first step. The true value of multi-omics analysis lies in the data interpretation—translating terabytes of raw information into actionable biological meaning. That's where our customized bioinformatics solutions come in.
Our expert team—comprising molecular biologists, statisticians, and data scientists—works side-by-side with you to tailor every analytical step, ensuring your multi-omics data analysis aligns with your biological hypothesis, study design, and publication goals.
Why Personalized Bioinformatics Matters
Standardized workflows may overlook subtle regulatory events or system-wide interactions. CD Genomics offers fully customized pipelines to:
- Integrate heterogeneous datasets (e.g., RNA-seq + ChIP-seq + LC-MS)
- Align omics data with public databases (TCGA, GEO, ENCODE)
- Construct regulatory and metabolic networks
- Uncover driver genes, epigenetic switches, and therapeutic targets
- Correlate molecular signatures with clinical phenotypes or treatment response
Our approach balances biological relevance and statistical rigor—providing results that are reproducible, insightful, and ready for peer review.
Core Analytical Capabilities
| Analysis Type | Key Deliverables |
|---|---|
| Differential Expression / Abundance | Volcano plots, heatmaps, fold-change tables |
| Multi-Omics Integration | MOFA, correlation matrices, cluster analysis |
| Pathway & GO Enrichment | KEGG, Reactome, STRING, GSEA |
| Network Construction | TF-gene networks, protein interaction maps |
| Methylation & Epigenetic Profiling | DMRs, chromatin accessibility maps (ATAC-seq) |
| Biomarker Discovery | Predictive markers linked to disease, traits, or response |
| Public Database Mining | GEO/TCGA data re-annotation and co-analysis |
| Clinical Association Mapping | Survival analysis, immune infiltration, risk scoring |
Workflow

Why Choose CD Genomics
A Trusted Partner for Multi-Omics Discovery
Advanced Technological Infrastructure
- Sequencing Platforms: Illumina NovaSeq, PacBio Sequel II, MGI DNBSEQ, and Oxford Nanopore
- Multi-Omics Ready: From WGS and RNA-seq to ChIP-seq, ATAC-seq, LC-MS/MS, and RRBS
- Automation & Throughput: High-volume capacity for large-scale omics studies
Expert Bioinformatics & Systems Biology Team
- Cross-functional specialists in oncology, microbiology, plant science, and data science
- Customized pipelines for multi-omics data integration, public database mining (TCGA, GEO), and pathway interpretation
- Publication-grade reporting with visualizations and regulatory insights
Flexible, End-to-End Service Ecosystem
- Support for single-omics and multi-omics projects across any stage
- One-stop workflow: sample QC → library prep → sequencing → data QC → integration → report
- Co-development of data strategies for novel targets, translational research, and publication planning
High-Impact Data Interpretation
- Incorporate clinical metadata, experimental controls, and phenotype correlations
- Use of advanced tools: MOFA, DESeq2, STRING, KEGG, Cytoscape, and machine learning algorithms
- Standardized QC metrics and reproducibility frameworks built into every project
Researcher-Oriented Support
- Bilingual project managers and PhD consultants available throughout the project
- Assistance with SCI manuscript preparation, journal submission, and peer review responses
- Long-term partnerships with CROs, academic labs, and pharmaceutical innovators
Downloadable Poster Showcase
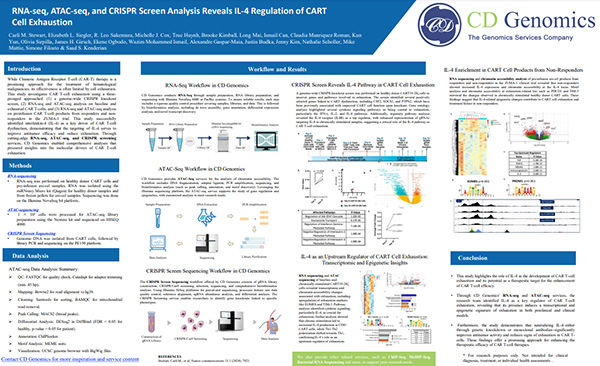
RNA-seq, ATAC-seq, and CRISPR Screening
Powering CAR T-Cell Research through Multi-Omics
Discover how integrated transcriptomic, epigenomic, and functional screening data reveals T-cell exhaustion mechanisms.
ChIP-seq and RNA-seq Integration
Decoding Mechanisms in Pancreatic Cancer Stem Cells
Explore how dual-omics profiling identifies epigenetic regulators and transcriptional shifts in CSC populations.

Sample Requirements
Streamlined Process. High-Quality Results.
We accept the following sample types for multi-omics projects:
- Genomics: Genomic DNA (≥1 μg, OD260/280 = 1.8–2.0)
- Transcriptomics: Total RNA (≥2 μg, RIN ≥7)
- Epigenomics: High-quality DNA or chromatin extracts
- Proteomics: Cell/tissue lysates or purified proteins (≥100 μg)
- Metabolomics: Biofluids, tissues, microbial pellets (flash-frozen, ≥100 mg or 200 μL)
Please consult with our technical team for species-specific requirements or custom formats.
Multi-Omics Service FAQs
Q1: What omics types can be integrated in one project?
We support flexible combinations of genomics, transcriptomics, epigenomics, proteomics, and metabolomics. You can choose 2–5 layers based on your biological question and project goals.
Q2: Can I provide my own sequencing data for integration?
Yes. We accept external datasets in standard formats and can integrate them with newly generated omics data or public databases (e.g., TCGA, GEO) as part of a customized analysis plan.
Q3: What kinds of research questions can multi-omics help answer?
Multi-omics analysis is ideal for uncovering regulatory mechanisms, identifying biomarkers, profiling disease subtypes, understanding stress or immune responses, and mapping host–microbiome interactions.
Q4: What deliverables will I receive?
You will receive a full report with annotated data tables, interactive plots, pathway diagrams, network maps, and publication-ready visual figures. Both raw and processed data files are included.
Q5: How do you ensure data quality and reproducibility?
We apply stringent quality control at every stage—sample assessment, sequencing, data preprocessing, and statistical modeling. Cross-validation across omics layers improves reliability and reduces noise.
Q6: Can you assist with manuscript preparation or downstream analysis?
Yes. We offer optional support for manuscript writing, figure formatting, and peer review responses. We also provide extended data mining and interpretation services upon request.
Q7: How should I get started with a multi-omics project?
Simply contact us with your project goals and available sample types. Our scientific team will help design the optimal omics strategy and guide you through sample submission and workflow planning.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
The HLA Class I Immunopeptidomes of AAV Capsid Proteins
Journal: Frontiers in Immunology
Year: 2023
Complete Genome Sequence of Bifidobacterium adolescentis Strain SD-VA1
Journal: Microbiology Resource Announcements
Year: 2020
Characterization of Salmonella from Retail Pork Using Whole Genome Sequencing
Journal: Frontiers in Microbiology
Year: 2022
DNA Methylation Predicts Recombination in Rice Using Whole-Genome Bisulfite Sequencing
Journal: Journal of Integrative Plant Biology
Year: 2023
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines

