Microsatellite Development
CD Genomics now offers a next generation sequencing based method to develop microsatellite markers for your species of interest, to benefit the scientific community.
The Introduction of Microsatellite Development
Microsatellites, also referred to as SSR, consist of a repeated motif with one to six Nucleotides, and they are ubiquitous in most eukaryotic genomes. Variations in the number of repetitions generate different alleles. SSR has high polymorphism, genomic specificity, abundance, and codominance. This makes them appropriate molecular markers for molecular-assisted breeding, molecular phylogenetics, and population genetics.
Next-generation sequencing (NGS) is recently used to develop microsatellite markers. Traditional methods depending on capillary sequencing are time-consuming and complex. Compared to traditional methods, NGS has the advantages of high-throughput, massively increasing output and low-cost. It can sufficiently analyze even non-model organisms. We utilized illumina NGS technology to produce millions of short fragment reads, which were screened with bioinformatics toolsets to identify primers that amplify polymorphic microsatellite loci.
The general workflow for microsatellite development is outlined below.
- DNA library preparation and shotgun sequencing with Illumina platform.
- Analysis of sequencing results with bioinformatics tools and identification of potential microsatellite loci for primer development.
- Synthesis of primers and selection of microsatellite loci by testing on several individuals to assess amplification and polymorphism.
- Analysis of individuals you would like us to genotype with only those selected primer pairs which amplified polymorphic loci.
We would screen at least 8 samples across up to 60 loci, attempt PCR optimization for all loci, finally we will do our best to identify 10 polymorphic loci, but we cannot guarantee because some population don't have 'normal' level of polymorphism.
Advantages of Microsatellite Development
The application of High-Throughput Sequencing (HTS) in microsatellite development considerably elevates its efficiency and precision, offering several notable advantages over traditional methods:
- Massive data acquisition: HTS can generate a considerable volume of genomic sequence data in a short period of time, significantly curtailing the timeline for microsatellite development.
- Automation: Sequencing to data analysis, a host of processes can be automated, reducing the need for manual intervention.
- Comprehensive coverage: With the potential to cover the major portion of the genome, including non-coding and low complexity regions, HTS increases the comprehensiveness of microsatellite identification.
- High resolution: HTS enhances accuracy in identifying the repeat units and amounts of microsatellites, thereby reducing the chances of misidentification.
- Reduced single-instance costs: Despite the higher initial equipment costs, progressive developments in technology have continually reduced sequencing costs on a per-instance basis, making large-scale microsatellite development more economical.
- Parallel processing: HTS allows for simultaneous processing of multiple samples, further reducing costs per unit sample.
- Possibility for multi-layered analysis: HTS offers not just microsatellite information, but can also be employed for other genomic analyses (like SNP detection, gene expression analysis, etc.), providing data support for comprehensive research.
- Discovery of New Microsatellites: Compared to traditional methods, HTS can identify more new microsatellite markers, thereby expanding the microsatellite marker library.
Application of Microsatellite Development
Studying Genetic Diversity and Population Structure: By leveraging high-throughput sequencing for the identification and development of numerous microsatellite markers, it is possible to perform comprehensive analyses examining the genetic structure of populations, gene flow, and genetic variation. This process can provide crucial insights into population dynamics as well as evolutionary processes.
Mapping Genomic Sequences and Locating Quantitative Trait Loci (QTL): Microsatellite markers play a vital role in the construction of genetic linkage maps as well as the localization of Quantitative Trait Loci (QTL). The precision and density of the markers, enhanced by high-throughput sequencing, have raised the bar in terms of the resolution achievable in genomic mapping.
Species Identification and Taxonomic Classification: Microsatellite markers developed using high-throughput technology can be effectively utilized in efforts related to species identification and taxonomic research. They assist in the recognition of genetic discrepancies between distinct species or populations, thereby substantiating efforts in the conservation and administration of biological diversity.
Research in Evolutionary Biology: Microsatellite markers, obtainable through high-throughput sequencing, can be instrumental in conducting evolutionary studies across various species or population groups. They can disclose the evolutionary history and phylogenetic relations of study subjects, thereby enriching our understanding of mechanisms behind biological evolution.
Agricultural and Breeding Applications: Within the realms of agriculture and breeding, microsatellite markers developed using high-throughput sequencing can be employed for breed identity confirmation, selection in breeding programs, and implementing genetic improvements in both plant and animal species. This enhances the efficacy and accuracy of agricultural and breeding efforts.
Microsatellite Development Workflow
The implementation of Next-Generation Sequencing (NGS) technology in the cultivation of microsatellites has markedly bolstered their efficiency, precision, and cost-effectiveness. Through a systematic progression that encompasses sample preparation, DNA sequencing, data processing, identification of microsatellites, primer design, validation of the primer, and testing of polymorphism, we can swiftly and effectively foster the development of microsatellite markers.
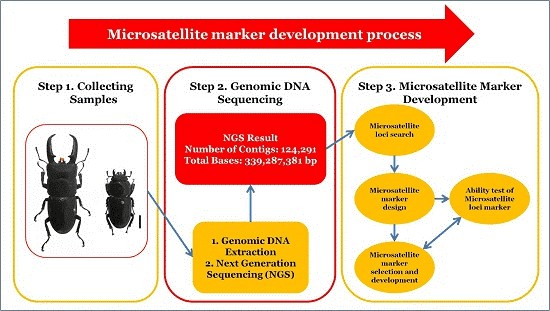
Service Specification
Sample Requirements
|
|
Sequencing Strategies
|
|
Data Analysis
|
Analysis Pipeline
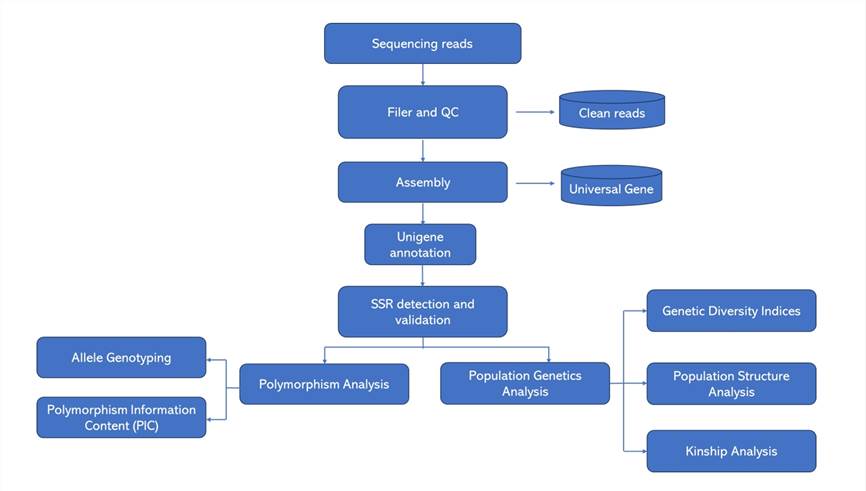
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
CD Genomics is dedicated to provide you with ready-to-use guaranteed, tested, polymorphic SSRs, and we have no legal right to any of your SSR markers. If you have additional requirements or questions, please feel free to contact us.
Reference:
-
Feng S, He R, Lu J, et al. Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Frontiers in Genetics. 2016, 7:113.
Demo Results
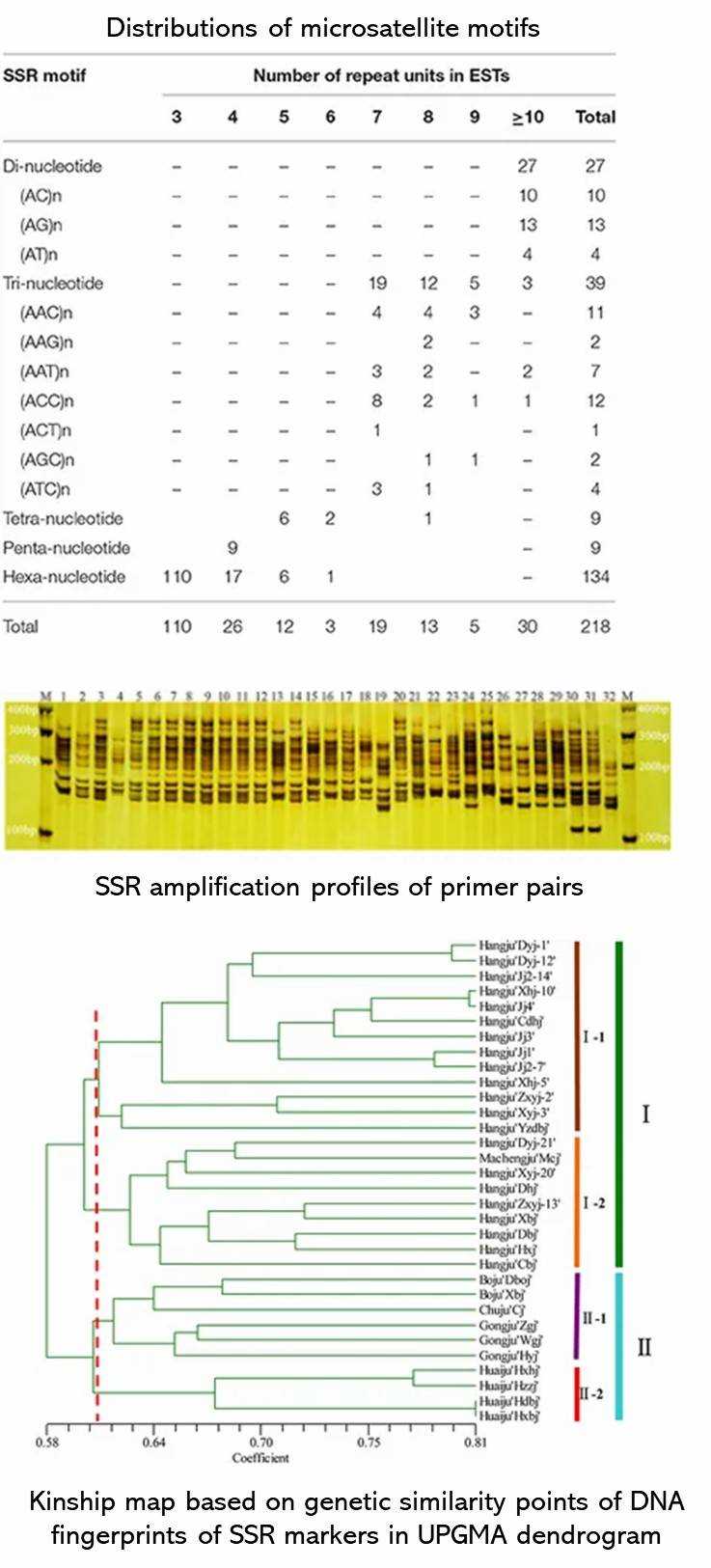 (Feng et al., 2016)
(Feng et al., 2016)
Microsatellite Development FAQs
1. Why are microsatellites important in genetic research?
Microsatellites hold substantial value as genetic markers within a range of genetic investigations, given their high variability, co-dominant inheritance, and prolific presence within genomes. They are frequently employed in the studies of population genetics, evolutionary biology, forensic examinations, paternity testing, and linkage mapping.
2. How are microsatellites developed?
The derivation of microsatellites typically begins with the high-throughput sequencing of genomic DNA, progressing towards bioinformatic scrutiny to identify repeating sequences. Subsequent to this, sequences are screened for microsatellite loci suitability, based on determinant factors such as the length of repeat motifs, quantity of repeats, and characteristics of flanking regions.
3. What factors should be considered during microsatellite primer design?
Designing microsatellite primers entails the scrupulous consideration of multiple important parameters. These include the length of the primers, which usually lies within 18 to 25 nucleotides, the melting temperature (Tm), the content of Guanine-Cytosine pairs (GC content), and the specificity towards the target. Together, these factors help to ensure the successful amplification of the target microsatellite locus through Polymerase Chain Reaction (PCR).
4. How are microsatellite markers validated?
The validation of microsatellite markers can be accomplished through PCR amplification focused on the desired loci, with generated primers which are designed specifically for the task. This step is then succeeded by electrophoresis analyses which examine and confirm the expected sizes of the amplicons. A subsequent scrutiny for polymorphism is done to check the disparities of microsatellite alleles across various organisms or populations.
5. What are some challenges associated with microsatellite development and application?
The development process of microsatellites can encounter certain challenges such as locating suitable loci that demonstrate a high degree of polymorphism, and mitigating the risk of genotyping errors that could result from stutter bands or allele dropout. Furthermore, challenges of a technical nature can arise in relation to the transferability across species and the development of multiplex PCR assays.
Microsatellite Development Case Studies
Development of Novel SSR Markers for Flax (Linum usitatissimum L.) Using Reduced-Representation Genome Sequencing
Journal: Fronts Plant Sci
Impact factor: 4.298
Published: 13 January 2017
Background
Flax breeding in northeastern China faces challenges in developing adaptable varieties with improved yield and quality. Marker-assisted selection (MAS) offers a solution, with recent advancements in SSR marker discovery showing promise. Utilizing next-generation sequencing methods, numerous SSR markers have been rapidly identified, aiding breeding programs. The Illumina sequencing platform, chosen for its high throughput and accuracy, has facilitated the systematic identification of SSR markers for immediate use in flax breeding.
Methods
- 48 cultivars/accessions
- DNA preparation
- Reduced Representation Genome Sequencing (RRGs)
- Illumina sequencing platform
- Shotgun sequencing
- Identification of SSRs
- SSR primer pair design
- Genetic diversity assay
Results
In the flax genome, the authors have identified 1,720 Simple Sequence Repeat (SSR) loci, of which 1,574 are novel findings. The SSR motif types predominantly consist of trinucleotide (56.1%) and dinucleotide (35.23%) repeats, per our data outlined in Table 1.
Table 1. Frequencies of different SSR repeat motif types.

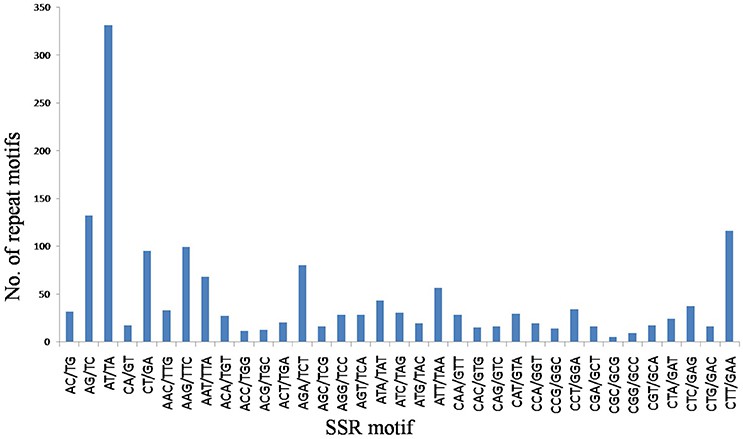 Fig 1. Numbers of dinucleotide and trinucleotide SSRs classified based on their motifs.
Fig 1. Numbers of dinucleotide and trinucleotide SSRs classified based on their motifs.
Using 62 primer pairs, polymorphism identification was conducted on 48 flax varieties from northeastern China, revealing their classification primarily into fiber and oil types (Fig 2). Remarkably, within the fiber type, cultivars "NEW1" and "Venus" exhibited significantly distinct genetic backgrounds. Similarly, the genetic background of "A0529" differed from other oil types. Consequently, these three flax cultivars hold significant implications for flax breeding endeavors.
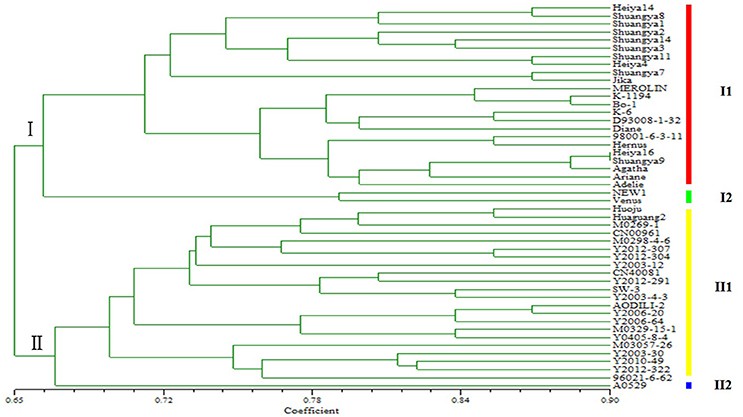 Fig 2. Genetic diversity of 48 flax cultivars/accessions based on SSR markers.
Fig 2. Genetic diversity of 48 flax cultivars/accessions based on SSR markers.
Conclusion
The authors developed 1574 novel SSRs in flax using reduced representation genome sequencing. Among these, 62 SSR sites were selected for primer design to assess genetic diversity in 48 flax varieties. Results showed clear differentiation between fiber and linseed flax varieties. These SSRs will be crucial for genetic analysis and mapping in flax and other crops.
Reference:
- Wu J, Zhao Q, Wu G, et al. Development of novel SSR markers for flax (Linum usitatissimum L.) using reduced-representation genome sequencing. Frontiers in plant science. 2017(7):2018.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Fungi: friends or foes—an outreach science initiative for the collection of airborne fungal spores by high school students
Journal: Journal of Microbiology and Biology Education
Year: 2024
Small but significant genetic differentiation among populations of Phyllachora maydis in the midwestern United States revealed by microsatellite (SSR) markers
Journal: bioRxiv
Year: 2023
The genetic legacy of fragmentation and overexploitation in the threatened medicinal African pepper-bark tree, Warburgia salutaris
Journal: Scientific Reports
Year: 2020
Evaluation of Plasma Biomarkers for A/T/N Classification of Alzheimer Disease Among Adults of Caribbean Hispanic Ethnicity
Journal: JAMA Network Open
Year: 2023
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
