CRISPR Screen Sequencing
Introduction to CRISPR Screen Sequencing
Effective CRISPR screening begins with a meticulously designed library of single guide RNAs (sgRNAs) targeting specific genes or loci. The design process involves selecting appropriate sequences, synthesizing the sgRNAs, and subsequently cloning them into vectors or transcribing them into RNA for cell transfection. Screening is then conducted using either positive or negative selection methodologies. Post-transfection, cells are physically sorted into distinct populations, followed by PCR amplification and high-throughput parallel sequencing. Data analysis subsequently identifies the sgRNAs and corresponding target genes of interest.
Utilizing CRISPR screening technology for high-throughput assays facilitates the generation of extensive mutant cell libraries, which can be subjected to various external conditions to isolate mutant cell subsets. High-throughput sequencing and bioinformatics analyses further elucidate the relationships between phenotypes and genotypes. This CRISPR/Cas9-based high-throughput screening offers significant advantages over RNA interference (RNAi) screening techniques, including overcoming issues related to low transfection efficiency and the limited ability to suppress gene expression at the mRNA level.
In the era of precision medicine, CRISPR screening holds tremendous scientific value and offers vast potential for research applications. This method provides a robust, simple, and programmable approach to large-scale gene editing—often referred to as genome-wide or high-throughput gene editing—which has demonstrated substantial efficacy, particularly in mammalian research.
To address the emerging needs of research communities, CD Genomics has developed an affordable, reliable, and high-throughput strategy for CRISPR Screen sequencing by harnessing amplicon based next-generation sequencing. Our CRISPR Screen sequencing service can give you direct and detailed information about sgRNA and targeted gene analysis and functional enrichment analysis, to help researchers study genes functions in a high-throughput fashion.
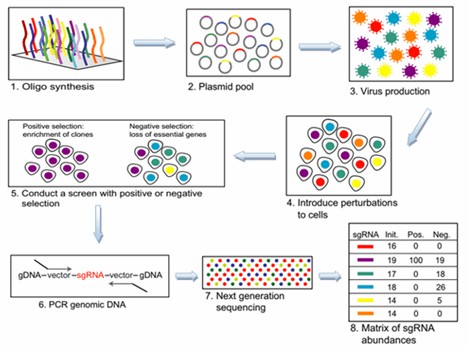 Figure 1. Overview of CRISPR Screening.
Figure 1. Overview of CRISPR Screening.
Advantages of Our CRISPR Screen Sequencing Service
- Efficient Screening: It edits multiple genes simultaneously, allowing for effective screening and evaluation of gene functions.
- Comprehensive Assessment: Provides a systematic approach to assessing gene functions within cells or organisms.
- Precise Genetic Modifications: Targets specific genomic areas to achieve accurate gene edits such as knockouts or mutations.
- Rapid Data Interpretation: Uses advanced sequencing technology for swift and accurate data analysis.
- Versatile Applications: Widely applied in discovering drug targets, establishing disease models, and advancing therapeutic development.
- Streamlined Processes: Enhances efficiency and reduces costs compared to conventional methods, speeding up research endeavors.
- Extensive multiplexing flexibility and high-throughput sequencing, enables for quantifying and comparing the frequencies of sgRNAs.
- Cost-effective, and highly sensitive detection levels without bias.
- No need for laborious and time-consuming cloning steps.
- Dedicated support from specialized Ph.D.-level scientists.
Applications of CRISPR Screen Sequencing
- Gene Function Studies: CRISPR Screen Sequencing allows researchers to systematically explore the functions of genes in cells or organisms, particularly their roles in complex biological processes and disease development.
- Drug Target Discovery: Through large-scale screening and analysis of genomic edits, potential drug targets can be discovered and evaluated for their roles in disease treatment.
- Disease Model Establishment: Based on CRISPR Screen Sequencing results, more accurate disease models can be established for studying disease mechanisms and screening therapeutic approaches.
CRISPR Screen Sequencing Workflow
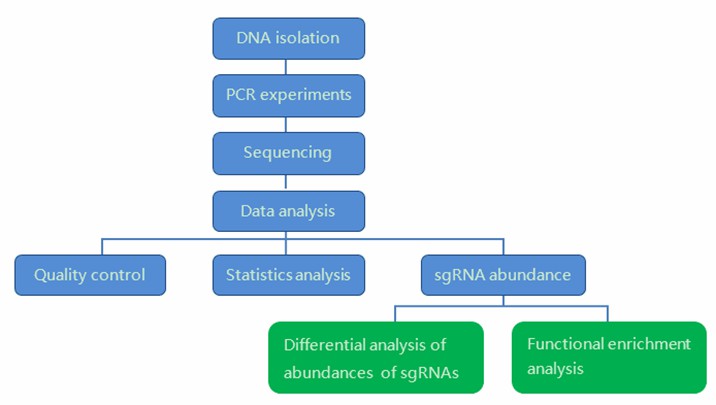
Our highly experienced expert team executes quality management following every procedure to ensure comprehensive and accurate results. Our CRISPR Screen sequencing workflow is outlined below, including DNA isolation, PCR experiments, sequencing, and bioinformatics analysis. Our CRISPR Screen sequencing also can help researchers to identify which gene knockouts yielded phenotypes relevant to the Screen.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in CRISPR Screen Sequencing for your writing (customization)
CD Genomics offers services for identifying sgRNA sequences and target genes of interest, followed by deep sequencing, at highly competitive prices. Our capabilities allow for the simultaneous sequencing of hundreds of samples. We utilize the MAGeCK tool for the analysis of sgRNA abundance and differential expression between groups. Should you have any additional requirements or inquiries, please do not hesitate to reach out to us.
References
- Ella H, et al. Genetic Screens and functional genomics using CRISPRCas9 technology. FEBS Journal, 2015, 1383-1393.
- Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature, 2014, 509(2):487-491.
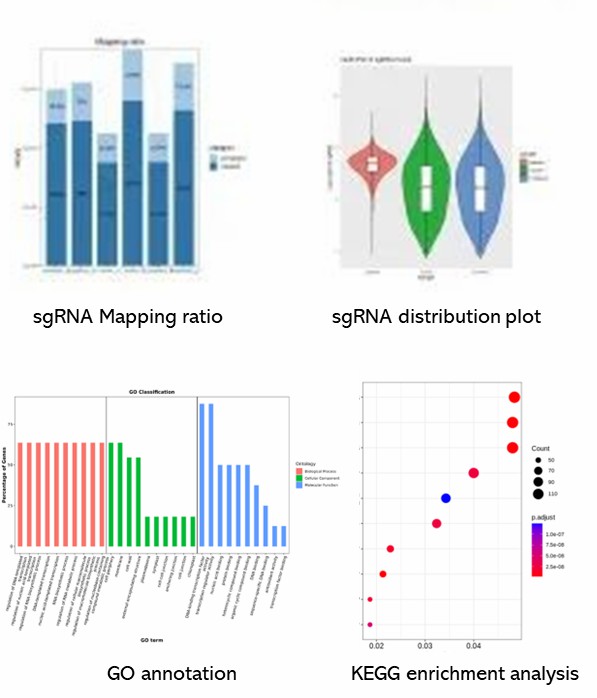
Demo Results
Partial results are shown below:
CRISPR Screen Seq FAQs
1. How to design sgRNA libraries for whole-genome or partial-genome sequences?
The design of sgRNA libraries for whole-genome or partial-genome sequencing is a critical aspect of effectively utilizing CRISPR-SCREEN technology. Achieving high editing efficiency, minimized off-target effects, and ensuring frameshift mutations upon gene knockout necessitate a comprehensive evaluation of multiple factors. The following key considerations are essential for optimal sgRNA design:
- Analysis of GC content;
- Selection of cutting positions;
- Analysis of secondary structures in the sequence;
- Assessment of gRNA specificity;
- Avoidance of sequences in the gRNA (excluding PAM) with four or more consecutive T nucleotides in the target sequence;
- Prediction of the gRNA sequence's efficiency score;
- Evaluation of potential off-target effects and mismatches in the gRNA sequence.
2. What types of CRISPR screens can be performed?
- Positive Selection Screens: Identify genes whose disruption confers a growth advantage or resistance under specific conditions.
- Negative Selection Screens: Identify genes whose disruption leads to a loss of viability or sensitivity to specific treatments.
- Pooled Screens: Utilize a mixed population of cells transduced with different sgRNAs, allowing high-throughput analysis.
- Arrayed Screens: Each well or sample contains cells transduced with a single sgRNA, facilitating detailed phenotypic analysis.
3. How do you ensure minimal off-target effects in CRISPR Screen Sequencing?
- sgRNA Design: Use bioinformatics tools to design sgRNAs with high specificity for target sites and minimal predicted off-target sites.
- Validation: Perform experiments to validate the specificity of selected sgRNAs.
- High-fidelity Cas9 Variants: Utilize Cas9 variants engineered for reduced off-target activity.
4. How do you analyze the data from CRISPR Screen Sequencing?
Data analysis involves several steps:
- Sequencing Data Quality Control: Check the quality of sequencing reads.
- Mapping Reads: Align the reads to the reference genome to identify sgRNA sequences.
- Hit Identification: Determine which sgRNAs are significantly enriched or depleted compared to controls.
- Functional Analysis: Interpret the biological significance of the identified hits, often using bioinformatics tools to analyze gene pathways and interactions.
CRISPR Screen Seq Case Studies
Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis
Journal: Journal of Experimental & Clinical Cancer Research
Impact factor: 12.568
Published: 15 January 2022
Background
Cancer metastasis involves tumor cell migration and invasion, requiring resistance to anoikis. The extracellular matrix (ECM) is crucial in this process. Using CRISPR/Cas9 knockout screening in ovarian cancer cells, PCMT1 was identified as essential for anoikis resistance. PCMT1 interacts with ECM protein LAMB3, activating pathways that aid in cell adhesion and invasion. Elevated PCMT1 in metastases highlights its potential as a therapeutic target.
Materials & Methods
Sample Preparation
- Patients
- Tissue samples
- RNA extraction
Sequencing
- Cell culture
- Genome-wide CRISPR/Cas9 screen
- Next-generation sequencing (NGS)
- IP-MS
- Quantitative real-time PCR
- RNAi Gene Enrichment Ranking (RIGER) analysis
- Western blot analysis
- Immunohistochemistry (IHC) analysis
- Statistical analyses
Results
A genome-wide CRISPR/Cas9 screen in ovarian cancer cells identified genes linked to anoikis resistance. Using the GeCKO v2 library, SKOV3 cells were screened under standard and ultralow-attachment conditions. Sequencing revealed 286 genes from negative selection and 122 from positive selection. Key pathways included protein repair and translational initiation, with known anoikis resistance genes like RAN, KIF11, and ECT2 also identified.
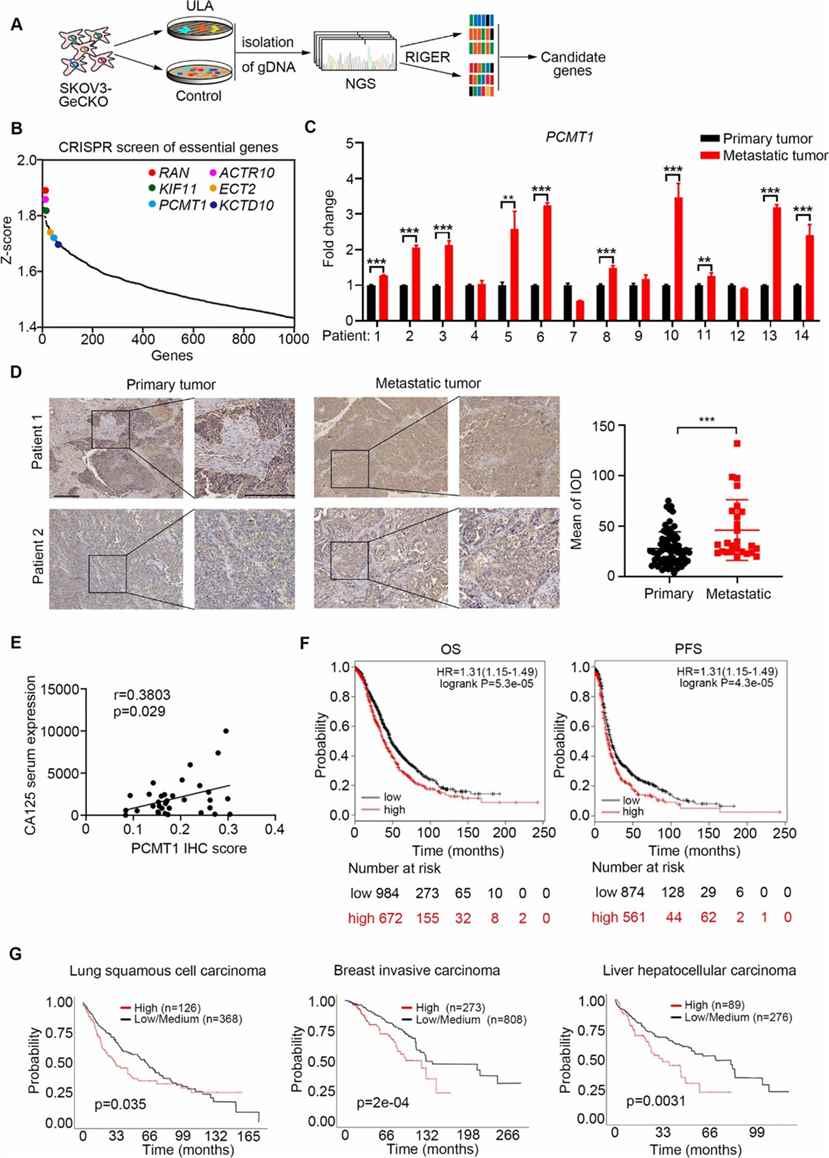 Fig 1. A pooled genome-wide CRISPR screen in an ovarian cancer cell metastasis model.
Fig 1. A pooled genome-wide CRISPR screen in an ovarian cancer cell metastasis model.
IP-MS identified that PCMT1 interacts with LAMB3. Among the 39 high-confidence PCMT1-interacting proteins, LAMB3 was linked to cell metastasis traits. Functional studies confirmed the interaction between PCMT1 and LAMB3, and silencing LAMB3 reduced cell spheroid formation and migration in PCMT1-overexpressing ovarian cancer cells, indicating that LAMB3 is a downstream target of PCMT1 in cancer metastasis.
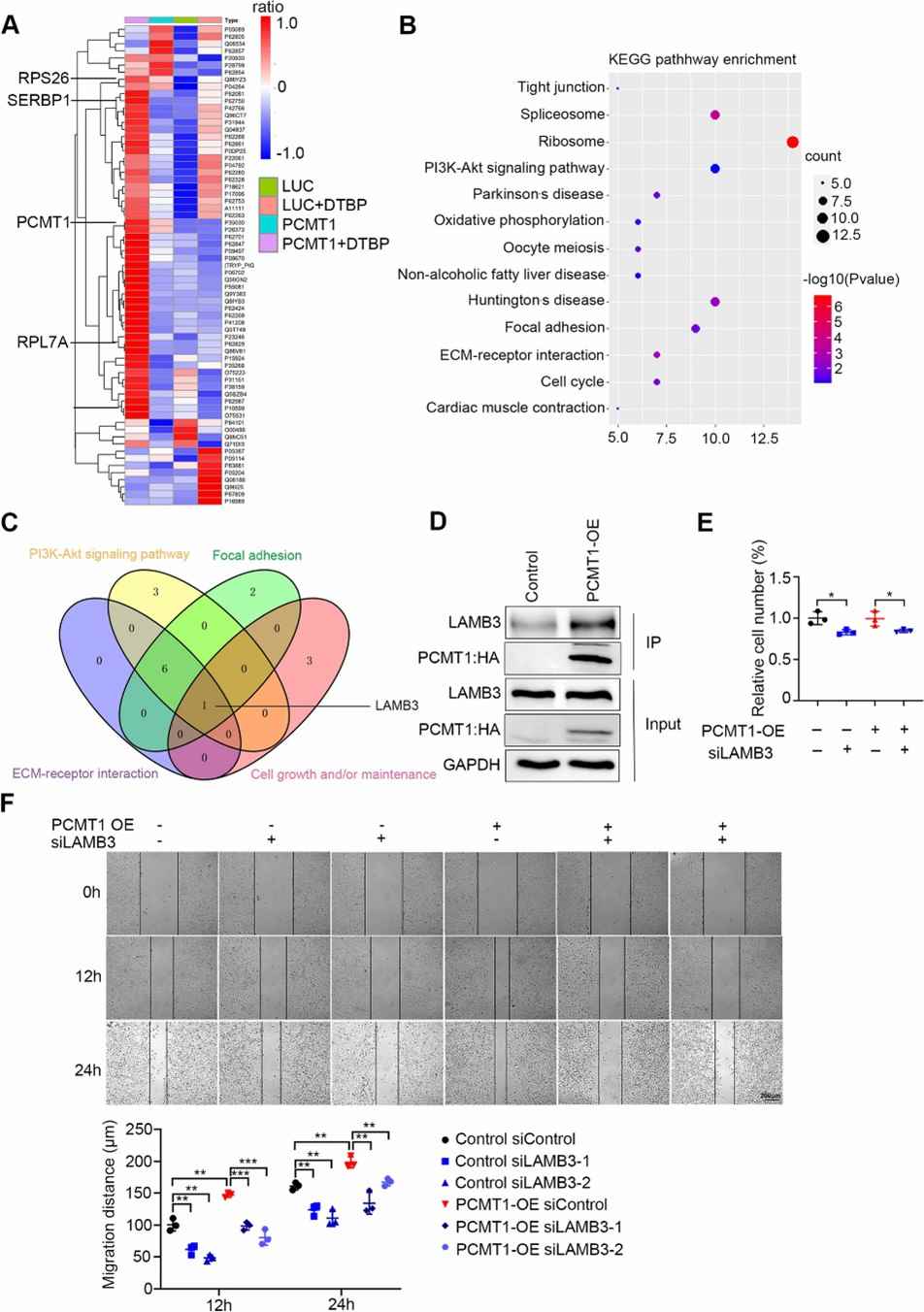 Fig 2. IP-MS indicates that PCMT1 interacts with LAMB3.
Fig 2. IP-MS indicates that PCMT1 interacts with LAMB3.
Conclusion
In summary, this study identifies PCMT1 as a key driver of anoikis resistance, enhancing cell adhesion, migration, and invasion. PCMT1 upregulates FAK-Src signaling, promotes metastasis, and correlates with metastatic stage in ovarian cancer. Elevated serum PCMT1 may indicate metastatic potential, and targeting PCMT1 with antibodies shows promise as a therapeutic strategy for ovarian cancer metastasis.
Reference
- Zhang J, Li Y, Liu H, et al. Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis. Journal of Experimental & Clinical Cancer Research, 2022, 41(1): 24.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
