Epigenomics Data Analysis Service: Unlocking the Secrets of Epigenetics
Epigenomics, a rapidly advancing field within genomics, delves into the intricate mechanisms that regulate gene expression through chemical modifications to DNA and associated proteins. These epigenetic changes have profound implications for various biological processes and can influence the development of diseases. As the understanding of epigenomics expands, so does the need for robust and comprehensive data analysis solutions. Any epigenomic investigation needs to carefully align the selection of the experimental protocols with the subsequent bioinformatics analysis and vice versa, as the effect sizes can be small and thereby escape detection if an integrative design is not well considered.
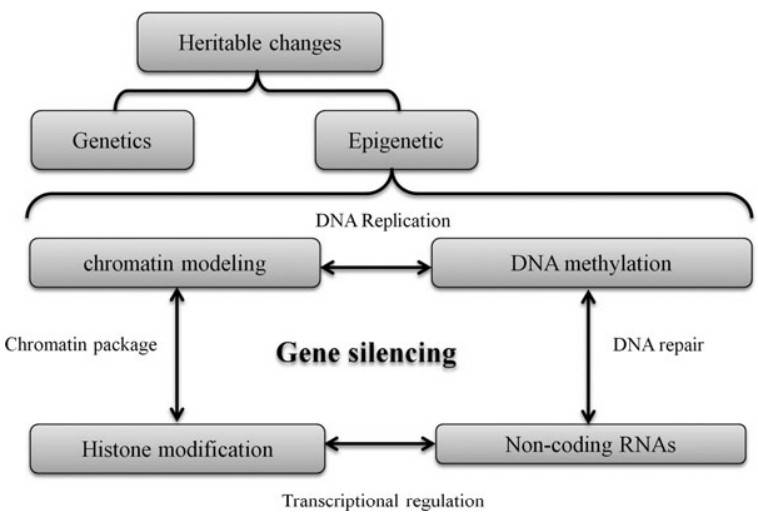 Interaction of different processes due to the epigenetics
Interaction of different processes due to the epigenetics
Bioinformatics Analysis of Epigenetics: Decoding the Language of the Epigenome
Bioinformatics plays a pivotal role in deciphering the language of the epigenome. Many tools and databases are available now that help to analyze epigenetic data, and these arebecoming increasingly important in solving the hidden mysteries of gene regulation. Bioinformatics tools and techniques help in various ways in the field of epigenetics, the major uses of which include understanding the biology of diseases caused by the epigenome so that therapies can be optimized, design of tools for analysis of the epigenetic data, and development of methods for analyzing and understanding the large epigenome datasets. Many approaches are used to analyze the epigenomic data which are helpful in aiding understanding the biology of diseases such as cancer as well as many heritable changes. Epigenomic data generated using experimental techniques such as chip sequencing can be further analyzed using bioinformatics tools and software and many databases have been created to enable retrieval of the epigenomic dataset of any specific tissue.
Bioinformatics or computational analysis have become useful approaches to understand epigenetic mechanisms and analyze the epigenomic data.Computational strategies have enabled various experimental techniques to evolve, which helps in generating a large amount of epigenetic data for processing and analysis; this requires suitable computational methods to process the data and control the quality of the epigenomic data. Predicted epigenomic data help in the understanding of the genomic distribution of epigenetic information.
Our Service
CD Genomics, a leading company in genomics research, offers an exceptional Epigenomics Data Analysis Service, empowering researchers and scientists to unravel the mysteries hidden within epigenetic data. We boast a team of highly skilled bioinformaticians who constantly optimize pipelines to deliver biologically meaningful results within a short turnaround time. By leveraging cutting-edge algorithms, computational tools, and machine learning techniques, CD Genomics enables researchers to extract valuable insights from their epigenomic datasets.
Chromatin studies (ChIP-Seq, ATAC-Seq)
Methylation studies (WGBS, RRBS, TBS, EPIC, MeDIP-Seq)
RNA-Seq
Peak Calling and Annotation
Exploratory analysis
Differential peak analysis
Transcription factor binding site analyses
Integrative analysis of multiple omics data
Data mining with machine learning techniques
Epigenomic Assays: Unveiling the Epigenetic Landscape
Epigenomic assays provide a window into the epigenetic landscape by profiling various aspects of chromatin structure, DNA methylation, and gene regulation. CD Genomics offers expertise in analyzing data generated from popular epigenomic assays, including:
Chromatin Studies (ChIP-Seq, ATAC-Seq): Understanding Chromatin Dynamics
Chromatin Immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) and Assay for Transposase-Accessible Chromatin followed by sequencing (ATAC-Seq) enable the investigation of DNA-protein interactions and chromatin accessibility, respectively. CD Genomics employs sophisticated analytical methods to process and interpret ChIP-Seq and ATAC-Seq data, providing valuable insights into chromatin dynamics and gene regulatory elements.
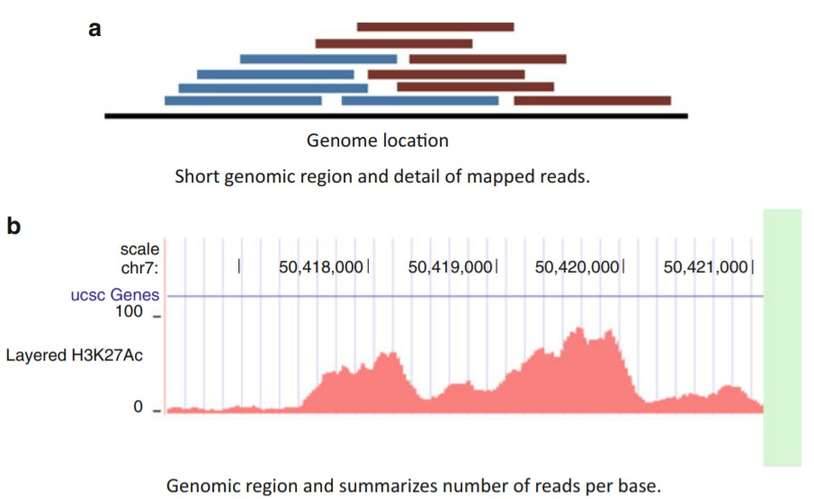 ChIP-seq mapped data
ChIP-seq mapped data
Methylation Studies (WGBS, RRBS, BSAS, EPIC, MeDIP-Seq): Decoding DNA Methylation Patterns
DNA methylation, a crucial epigenetic modification, plays a pivotal role in gene expression regulation. CD Genomics offers analysis services for various methylation studies, such as Whole Genome Bisulfite Sequencing (WGBS), Reduced Representation Bisulfite Sequencing (RRBS), Bisulfite Amplicon Sequencing (BSAS), EPIC arrays, and Methylated DNA Immunoprecipitation followed by sequencing (MeDIP-Seq). Through these approaches, researchers can gain insights into the DNA methylation landscape at single-base or methylated region resolution.
DNA Methylation Data Analysis: Deciphering the Methylome
CD Genomics excels in DNA methylation data analysis, starting from quality control and alignment of sequencing reads or normalization of array data. The identification of methylated sites allows for the discovery of larger regions of DNA methylation or differentially methylated regions (DMRs) between samples. CD Genomics provides comprehensive annotation of DMRs, facilitating their interpretation in the context of genes, regulatory elements, and other genomic features. Integration with gene expression data enables researchers to investigate the association between DNA methylation and gene expression, paving the way for the discovery of epigenetic biomarkers and the exploration of biological aging.
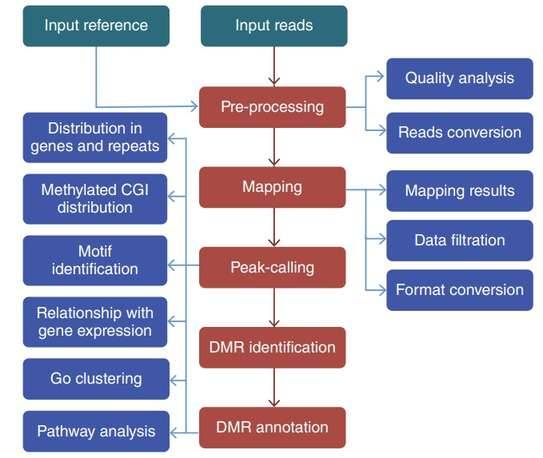 The workflow of DNA methylation analysis.
The workflow of DNA methylation analysis.
Transcription Factor Binding Site Analyses: Unraveling Transcriptional Regulation
Epigenomic data, particularly ChIP-seq and ATAC-seq, can shed light on transcription factor binding sites and their role in gene regulation. CD Genomics offers expertise in identifying transcription factor binding sites across the genome using ChIP-seq data. Through TF footprinting analysis, narrow drops in chromatin accessibility can be interpreted as protein binding sites, providing valuable information about the transcription factors involved. Coupling this information with RNA-seq data allows researchers to investigate the combined effects of transcription factors on gene expression in a high-throughput manner.
Peak Calling and Annotation: Identifying Regions of Interest
One of the fundamental steps in epigenomic data analysis is peak calling and annotation. CD Genomics utilizes advanced algorithms and peak caller tools to identify regions of interest that exhibit significant epigenetic signals. By accurately pinpointing these peaks, researchers can focus their analysis on genomic regions with potential functional significance.
Furthermore, CD Genomics provides comprehensive annotation services, enriching the identified peaks with relevant information. This includes associating peaks with nearby genes, regulatory elements, and binding motifs. The annotation of peaks enables researchers to perform gene set enrichment analyses and gain deeper insights into the downstream effects of epigenetic modifications.
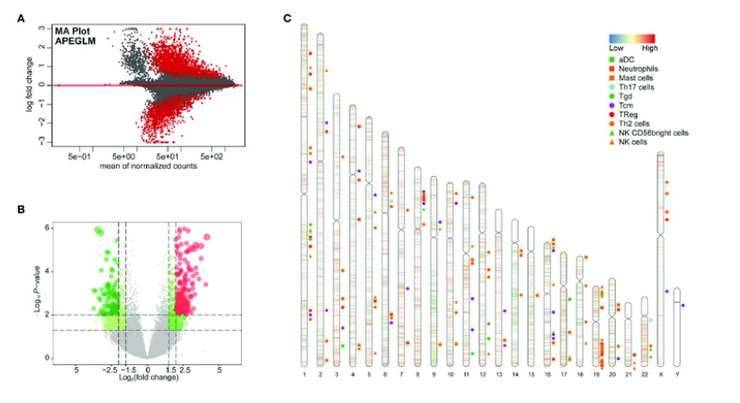 Differential peak analysis of TCGA-KIRC ATAC-seq data
Differential peak analysis of TCGA-KIRC ATAC-seq data
Exploratory Analysis: Visualizing and Interpreting Epigenomic Profiles
Exploratory analysis plays a vital role in understanding the characteristics and patterns within epigenomic data. CD Genomics employs powerful visualization techniques such as Principal Component Analysis (PCA), UMAP, and t-SNE to generate informative visual representations of epigenomic profiles. These visualizations help researchers identify similarities between biological replicates, detect distinct sample groups based on different tissues, treatments, or time points, and identify potential outlier samples requiring further investigation.
Differential Peak Analysis: Uncovering Epigenetic Differences
Comparing epigenomic profiles between different conditions is essential for understanding the dynamic nature of epigenetic regulation. CD Genomics specializes in differential peak analysis, enabling researchers to identify statistically significant differences in epigenetic signals between samples or experimental conditions. By quantifying the effect size and statistical significance, researchers can gain insights into the epigenetic alterations associated with specific biological contexts. Visualizations, such as volcano plots, aid in the interpretation of differential peak analysis results and facilitate the identification of regions with substantial changes in epigenetic states.
Integrating RNA-Seq and Epigenomic Data: Unveiling Gene Regulatory Networks
Integrating epigenetic information with transcriptomic data is crucial for understanding gene regulatory networks. Performing RNA-Seq and epigenomic sequencing, such as ChIP-seq or ATAC-seq, on the same samples enables integrative analysis, offering a comprehensive understanding of gene regulatory programs. CD Genomics specializes in RNA-Seq data analysis, providing researchers with a comprehensive understanding of gene expression and the interplay between epigenetic modifications and transcriptional activity. CD Genomics specializes in integrating these multi-omics datasets, allowing researchers to unravel regulatory connections between enhancers and their target genes, as well as transcription factors and their targets. By combining gene expression data with epigenomic information, researchers can uncover key regulatory mechanisms, identify master regulators, and gain insights into the functional consequences of epigenetic modifications on gene expression.
Integrative Analysis of Multiple Omics Data: Unifying the Epigenomic Landscape
Integrating multiple omics data, such as epigenomic, transcriptomic, and proteomic data, offers a holistic view of biological processes. CD Genomics excels in integrative analysis, employing sophisticated computational approaches to uncover connections between different omics layers and unveil novel insights into the complex interplay of epigenetic mechanisms in development and disease.
Bioinformatics Data Mining: Extracting Hidden Gems from Epigenomic Data
In the era of big data, bioinformatics data mining techniques are essential for extracting valuable information from large-scale epigenomic datasets. CD Genomics harnesses advanced data mining algorithms and approaches to identify novel patterns, correlations, and associations within epigenomic data. By applying machine learning and statistical techniques, CD Genomics can uncover hidden relationships between epigenetic features, identify epigenetic signatures associated with specific phenotypes or diseases, and predict the functional impact of epigenetic alterations.
CD Genomics: Your Trusted Partner in Epigenomics Data Analysis
As a leader in genomics research, CD Genomics is committed to providing high-quality and comprehensive epigenomics data analysis services. With our team of experienced bioinformaticians, cutting-edge technologies, and state-of-the-art analytical pipelines, we empower researchers and scientists to unlock the secrets of the epigenome and advance our understanding of gene regulation and disease mechanisms.
Whether you require epigenomic assay analysis, peak calling and annotation, differential peak analysis, transcription factor binding site analyses, DNA methylation data analysis, integrative analysis, or bioinformatics data mining, CD Genomics offers tailored solutions to meet your specific research needs. Our robust methodologies, rigorous quality control, and attention to detail ensure accurate and reliable results that pave the way for groundbreaking discoveries.
Advantages of CD Genomics' Epigenomics Data Analysis Service:
Expertise in Bioinformatics Analysis: Our team of skilled bioinformaticians has extensive experience in analyzing epigenomic data, ensuring accurate and reliable results.
Tailored Solutions: We offer customized analysis solutions to meet your specific research goals and requirements.
Cutting-Edge Technologies: We utilize advanced sequencing technologies for high-quality data generation, including ChIP-seq, ATAC-seq, WGBS, RRBS, and RNA-seq.
Comprehensive Service Offerings: Our services cover a wide range of epigenomic analysis types, enabling a holistic understanding of your data.
Rapid Turnaround Time: We optimize analysis pipelines to deliver biologically meaningful results within a short timeframe.
Collaborative Approach: We work closely with you, seeking your input and understanding your analysis requirements.
Manuscript Support: We assist in shaping the methods section of your manuscripts, ensuring accurate and transparent reporting of analysis methods and results.
Data Security and Confidentiality: We prioritize data security and adhere to strict measures to protect your data and ensure confidentiality.
Contact CD Genomics today to unlock the power of epigenomics and revolutionize your research!
References:
- Shiqiang Zhang et al,. Systematic Chromatin Accessibility Analysis Based on Different Immunological Subtypes of Clear Cell Renal. Cell Carcinoma Frontiers in Oncology 2021
- F. Marabita et al. Introduction to Data Types in Epigenomics,2015
- Zhanjiang (John) Liu, Yanghua He and Jiuzhou Song. Bioinformatics Analysis of Epigenetics. Bioinformatics in Aquaculture Principles and Methods. 2017
- Budhayash Gautam, Kavita Goswami, Neeti Sanan Mishra, Gulshan Wadhwa, and Satendra Singh. The Role of Bioinformatics in Epigenetics. Current trends in Bioinformatics: An Insight. 2018


 Sample Submission Guidelines
Sample Submission Guidelines
