5mC/5hmC Sequencing
CD Genomics offers advanced 5mC/5hmC sequencing services to uncover DNA methylation and hydroxymethylation patterns. Our high-resolution approach provides crucial insights into epigenetic regulation across genomes, supporting research in developmental biology, disease mechanisms, and beyond. Contact us to advance your epigenetics research today.
What is 5mC and 5hmC
DNA methylation and hydroxymethylation at the 5-position of cytosine (5mC) are essential epigenetic alterations that play vital roles in gene expression regulation and cell differentiation. DNA methylation typically represses gene transcription. Hydroxymethylation, on the contrary, activates gene expression or prompts DNA demethylation. 5mC groups introduced by DNA Methyl-Transferases can be iteratively oxidized to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) by the action of Tet Methylcytosine Dioxygenases. 5hmC is the most abundant component in vivo among the three 5mC oxidative derivatives and can be identified in almost all mammalian tissues and cells. 5mC and 5hmC genome-wide mapping uncover the genomic areas of these alterations, which is crucial for elucidating their mechanisms.
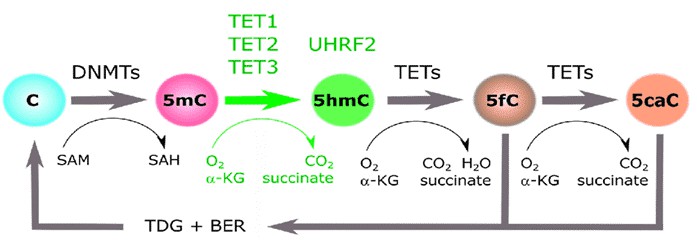 Figure 1. The cytosine methylation cycle (Ecsedi, 2018)
Figure 1. The cytosine methylation cycle (Ecsedi, 2018)
The Introduction of 5mC/5hmC Sequencing
Whole-genome bisulfite sequencing (WGBS) is considered the "gold standard" for methylation sequencing, primarily detecting the two forms of modification, 5mC and 5hmC. From this, several derivative methods have been developed to distinguish between 5mC and 5hmC. For instance, oxidation bisulfite sequencing (oxBS-seq) is used for 5mC-specific sequencing, and TET-assisted bisulfite sequencing (TAB-seq) is specific for detecting 5hmC. In recent years, 5hmC has emerged as a significant biomarker for cancer screening, demonstrating crucial clinical value.
Conventional bisulfite sequencing (BS-seq) does not differentiate between 5mC and 5hmC, presenting a mixed signal of the two modifications. Oxidation-bisulfite sequencing (oxBS-seq) oxidizes 5hmC to 5fC, subsequently converting it to the base U with bisulfite treatment, enabling precise detection of 5mC. Simultaneous BS-Seq and oxBS-Seq sequencing of the same sample can achieve single-base resolution detection of 5hmC, providing a hydroxymethylation-sensitive base resolution approach.
CD Genomics is a biotechnology company focusing on sequencing technology. With cutting-edge sequencing technology and in-depth bioinformatics analysis, we assist customers in mining genomics information. Our areas of expertise include genomics, transcriptomics, epigenetics, population genetics, microbiology, and many other fields. Our high-throughput 5mC/5hmC sequencing service utilizes multiple mature and stable platforms with high efficiency, simplicity, and accuracy to help your epigenetics research.
CD Genomics can provide genome-wide 5mC and 5hmC detection by following approaches:
1. Antibody-based immunoprecipitation and sequencing of hydroxymethylated DNA (hMeDIP-seq).
2. Oxidative bisulfite sequencing (oxBS-seq)
3. Whole-genome bisulfite/oxidative bisulfite sequencing (WGBS/oxWGBS-seq)
4. Reduced representative bisulfite oxidative bisulfite sequencing (RRBS/oxRRBS)
Advantages of 5mC/5hmC Sequencing
- High-Resolution Methylation Profiling: This methodology provides detailed mapping of DNA methylation (5mC) and hydroxymethylation (5hmC) throughout the genome, offering precise insights into methylation patterns not only at CpG sites but also beyond.
- Quantitative Measurement: It allows for the precise quantification of methylation levels in specific genomic regions such as promoters and CpG islands, granting comprehensive understanding of the mechanisms underlying epigenetic regulation.
- Whole-Genome Coverage: Encompassing the entire genome, this technique extends its reach to remote and non-coding regions, thus enabling a holistic grasp of methylation's involvement in gene regulation and broader epigenetic phenomena.
- High-throughput and Efficiency: Characterized by its capacity to efficiently manage large sample sizes, this approach is particularly adept at handling complex datasets and facilitating high-throughput analyses in a streamlined manner.
- Comparative Analysis: By supporting comparative evaluations of methylation profiles across multiple samples, it unravels variations correlated with diverse physiological states, diseases, or experimental settings, uncovering valuable insights into biological responses at the epigenetic level.
Applications of 5mC/5hmC Sequencing
- Disease Research and Diagnosis: Identifies methylation markers associated with diseases, potentially serving as early diagnostic or prognostic biomarkers in cancer and other disorders.
- Development and Differentiation: Investigates epigenetic mechanisms in biological development and differentiation processes, elucidating methylation's role in cell fate determination and tissue-specific gene expression.
- Environmental Response and Genetic Variation: Analyzes how environmental factors influence methylation patterns and explores the relationship between genetic variations and epigenetic changes, revealing insights into environmental adaptation and phenotypic plasticity.
- Drug Development and Therapeutic Strategies: Evaluates the impact of candidate drugs on methylation patterns, contributing to the discovery of new therapeutic targets and strategies.
- Agriculture and Plant Biology: Studies methylation patterns in plant genomes to understand their role in crop breeding, stress tolerance improvement, and agricultural productivity.
5mC/5hmC Sequencing Workflow
Our long-standing experience and advanced platforms allow us to provide a comprehensive service package from project consultation, experiment design, sequencing, to bioinformatics analysis. We strive to provide the most suitable scheme for your project.
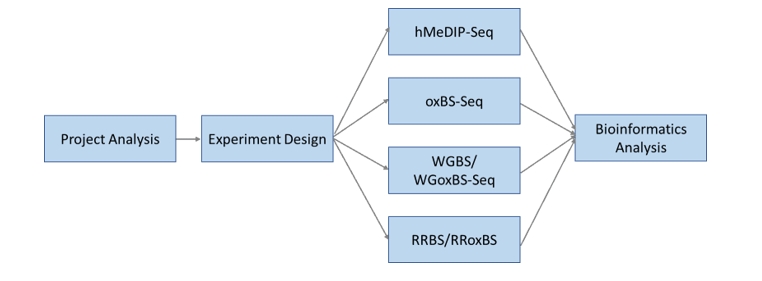
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
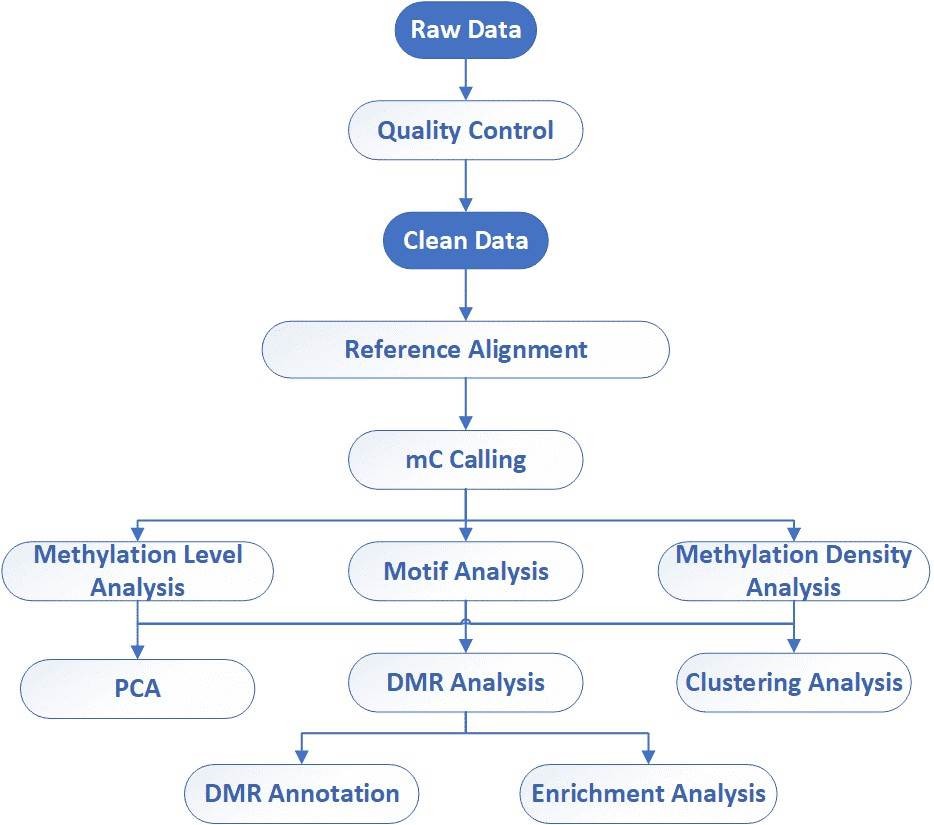
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in 5mC/5hmC Sequencing for your writing (customization)
Explore DNA methylation and hydroxymethylation with CD Genomics' 5mC/5hmC sequencing service. We provide high-resolution sequencing and comprehensive bioinformatics analysis to reveal epigenetic patterns across the genome. Contact us to unlock insights for your research.
References
- Ecsedi S, Rodríguez-Aguilera JR, Hernandez-Vargas H. 5-Hydroxymethylcytosine (5hmC), or how to identify your favorite cell. Epigenomes. 2018, 2(1):3.
- Kirschner K, Krueger F, Green AR, Chandra T. Multiplexing for Oxidative Bisulfite Sequencing (oxBS-seq). InDNA Methylation Protocols 2018. Humana Press.
Demo Results
Partial results are shown below:
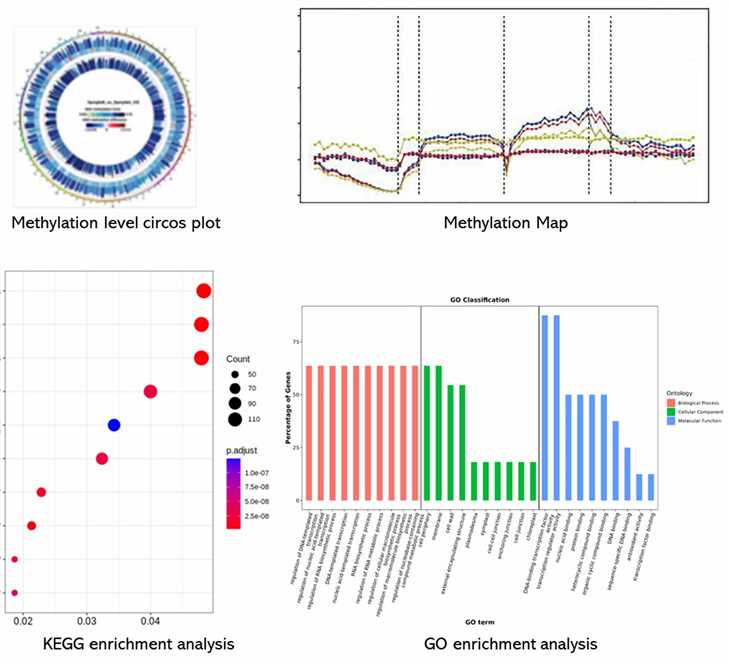
5mC/5hmC Seq FAQs
1. How does 5hmC influence gene transcription?
This epigenetic modification, 5-hydroxymethylcytosine (5hmC), plays a crucial role in gene transcription by modulating chromatin structure and orchestrating the recruitment of specific regulatory proteins. Its association with active transcriptional states hints at its capacity to facilitate gene expression processes. Notably, 5hmC disrupts DNA methylation dynamics, functioning as an intermediary in demethylation processes, thereby shaping gene regulatory networks.
2. Why is 5hmC significant?
Its role as a dynamic marker of DNA methylation alterations underscores its vital contribution to regulating critical biological processes like development, differentiation, and disease progression. Diverging from the repressive effects of 5-methylcytosine (5mC), 5hmC is attributed to activating transcriptional activities and regulatory functions within the genome. Unraveling the intricate patterns of 5hmC offers crucial insights into the epigenetic mechanisms governing normal physiological functions and pathological states, holding promise for diagnostic and therapeutic advancements in conditions such as cancer and neurological disorders.
3. How is 5hmC different from 5mC in terms of biological function?
In the realm of DNA methylation derivatives, a notable difference arises between 5hmC and 5mC. While both molecules partake in this epigenetic modification, 5hmC distinguishes itself through its affiliation with active transcription and involvement in the intricate processes of DNA demethylation. Notably, its influence on gene expression dynamics and developmental pathways stands in stark contrast to the traditional gene silencing role played by 5mC.
4. How can 5mC/5hmC sequencing contribute to personalized medicine?
Harnessing the power of 5mC/5hmC sequencing opens up avenues for uncovering epigenetic markers linked to disease susceptibility, prognosis, and treatment responsiveness. This innovative approach holds promise in tailoring personalized diagnostic and therapeutic interventions, thereby revolutionizing the landscape of precision medicine.
5mC/5hmC Seq Case Studies
Integrated analyses of multi-omics reveal global patterns of methylation and hydroxymethylation and screen the tumor suppressive roles of HADHB in colorectal cancer
Journal: Clinical epigenetics
Impact factor: 7.259
Published: 02 March 2018
Background
DNA methylation is a crucial epigenetic modification related to gene expression. 5mC and 5-hmC serve as two epigenetic markers involved in maintaining epigenetic reprogramming, with genome methylation potentially leading to cancer. The aim of this study is to elucidate the epigenetic mechanisms of DNA methylation and hydroxymethylation in colorectal cancer development. The authors employed MeDIP-seq and hMeDIP-seq to map the whole-genome DNA methylation and hydroxymethylation patterns of colorectal cancer tissues and corresponding normal tissues, along with transcriptome gene expression analysis (RNA-seq). Differential methylation regions (DMRs), differential hydroxymethylation regions (DhMRs), and differentially expressed gene regions (DEGs) were identified. Epigenetic biomarkers were selected through DMR, DhMR, and DEG analyses, followed by functional analysis for validation.
Materials & Methods
Sample Preparation
- Colorectal tumor samples
- Adjacent normal tissues
- DNA extraction
- RNA extraction
Sequencing
- RNA-seq
- MeDIP-seq
- hMeDIP-seq
- Quantitative PCR
- Gene Expression Level Analysis
- Differential Gene Expression Analysis
- Identification of DMR and DhMR
- Functional Enrichment Analysis of DMR and DhMR
- RNA interference analysis
- Western blot analysis
Results
The authors investigated the distinct whole-genome hydroxymethylation patterns that can serve as epigenetic biomarkers in normal colorectal tissue. By comparing colorectal tumor and normal tissues, a total of 59,249 DMRs, 187,172 DhMRs, and 948 DEGs were identified. Upon cross-referencing the genes from DMRs or DhMRs with DEGs, seven genes were selected as being aberrantly regulated by DNA methylation in tumors. Furthermore, high methylation of the HADHB gene was found to be correlated with its downregulation in colorectal cancer (CRC) transcription. Functional analysis of the tumor indicated that the HADHB gene reduced cancer cell migration and invasiveness. The results suggest that HADHB may act as a tumor suppressor gene (TSG).
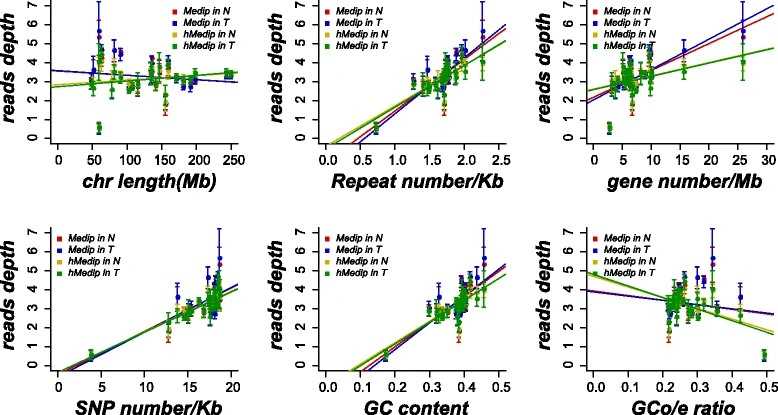 Fig. 1 Global patterns of methylome and hydroxymethylome.
Fig. 1 Global patterns of methylome and hydroxymethylome.
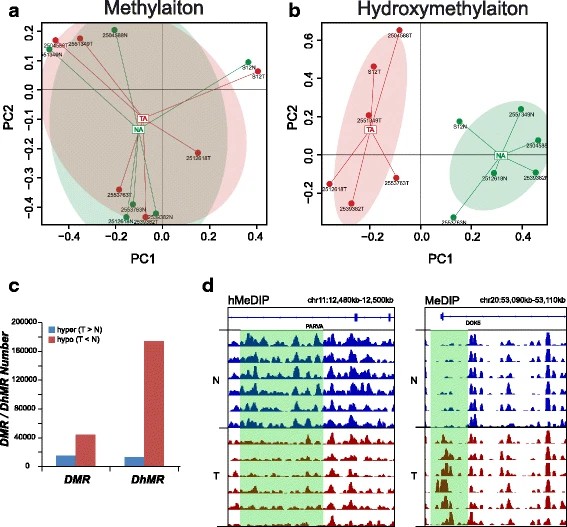 Fig. 2 Different distributions of methylation and hydroxymethylation.
Fig. 2 Different distributions of methylation and hydroxymethylation.
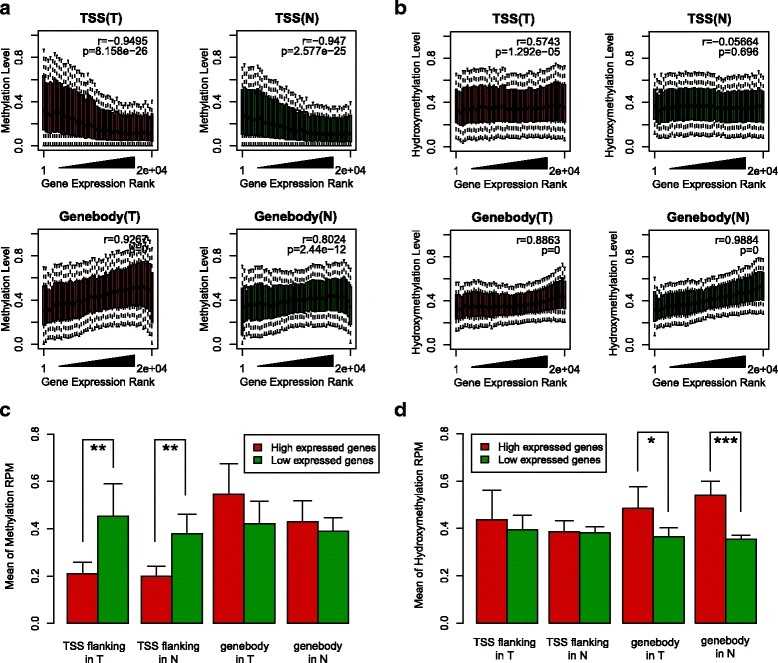 Fig. 3 Association between epigenomic modification and gene expression.
Fig. 3 Association between epigenomic modification and gene expression.
Conclusion
The study elucidates the genome-wide methylation and hydroxymethylation patterns in CRC. By employing multi-omics analyses, seven CRC-related genes were identified. Functional experiments supported that HADHB acts as a potential tumor suppressor gene, reducing the invasiveness and migration of tumor cells. The silencing of the HADHB gene may promote the onset and progression of colorectal cancer.
Reference
- Zhu Y, Lu H, Zhang D, et al. Integrated analyses of multi-omics reveal global patterns of methylation and hydroxymethylation and screen the tumor suppressive roles of HADHB in colorectal cancer. Clinical epigenetics. 2018, 10:1-3.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Restriction endonuclease cleavage of phage DNA enables resuscitation from Cas13-induced bacterial dormancy
Journal: Nature microbiology
Year: 2023
IL-4 drives exhaustion of CD8+ CART cells
Journal: Nature Communications
Year: 2024
High-Fat Diets Fed during Pregnancy Cause Changes to Pancreatic Tissue DNA Methylation and Protein Expression in the Offspring: A Multi-Omics Approach
Journal: International Journal of Molecular Sciences
Year: 2024
KMT2A associates with PHF5A-PHF14-HMG20A-RAI1 subcomplex in pancreatic cancer stem cells and epigenetically regulates their characteristics
Journal: Nature communications
Year: 2023
Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch
Journal: Science Advances
Year: 2024
Genomic imprinting-like monoallelic paternal expression determines sex of channel catfish
Journal: Science Advances
Year: 2022
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
