MassARRAY SNP Genotyping
CD Genomics offers a rapid and accurate custom SNP validation on the MassARRAY MALDI–TOF instrument provided by Agena Bioscience. Our MassARRAY SNP Genotyping system combines mass spectrometry, sensitive and robust chemistry, and advanced data analysis software to meet your SNP genotyping needs.
Introduction of MassARRAY SNP Genotyping
MassARRAY SNP Genotyping amplifies the DNA sequence across the SNP site by PCR, and then uses a single extension primer to amplify PCR products, ensuring that the primer extends only one base. The extended product was analyzed by TOF (time of flight) and the SNP was classified according to the molecular weight difference of base, and the allele frequency of the SNP site could also be calculated. MassARRAY SNP genotyping can carry out multiple SNP typing in one reaction. This technique is accurate and has a price advantage when analyzing dozens or hundreds of SNPs.
With different detection and software modules, the MassARRAY® iPLEX GOLD technology can realize the research or application of SNP genotyping, gene expression research, copy number variation, gene methylation analysis, pathogen typing and prenatal diagnosis on one platform. MassARRAY SNP Genotyping has a very good application prospect in the fields of biological science, medicine, agricultural research and so on. It has provided excellent solutions for scientists all over the world.
Advantages of MassARRAY SNP Genotyping
- Accurate and Automated: Nucleic acid detection by mass spectrometry enables readout by molecular mass, and using state-of-the-art technologies for automated, non-contact dispensing of reagents.
- Scalable and Flexible: We offer a variety of options for SNP, sample and assay throughput.
- High Multiplex Capability: Multiplex up to 30 SNP loci in a single pool.
- Fast Assay Design: A new assay can be designed within a few hours. In addition, a semi-automated workflow and unmodified oligos provide rapid turnaround time.
- Cost Effective: Multiplex analysis reduces per-sample cost, and scalable throughput optimizes batch and resource requirements.
Applications of MassARRAY SNP Genotyping
- Genotyping and mutation detection
- Ultrasensitive detection
- Methylation analysis
- Variety identification
- Functional Panel customization
- Pharmacogenomics research
- Biomarker validation
MassARRAY SNP Genotyping Workflow
The procedures for our MassARRAY SNP Genotyping is illustrated below.
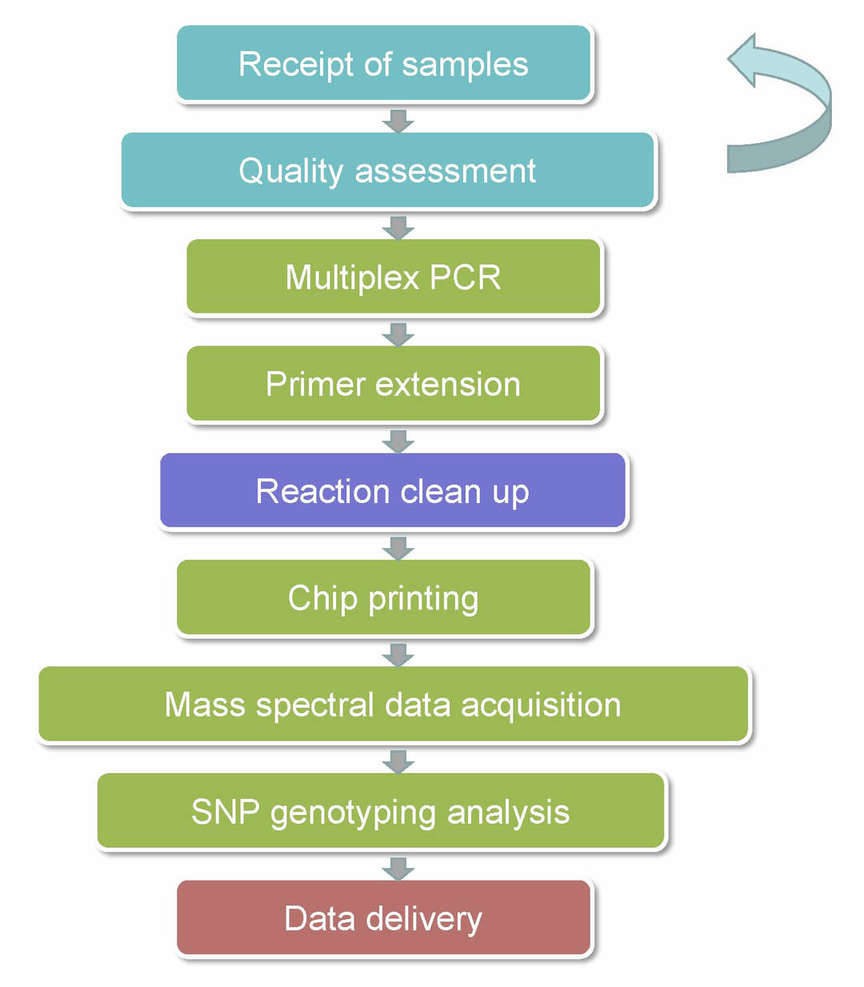
Service Specifications
Sample requirements
|
|
| Chip Printing | |
Data Analysis
|
Analysis Pipeline
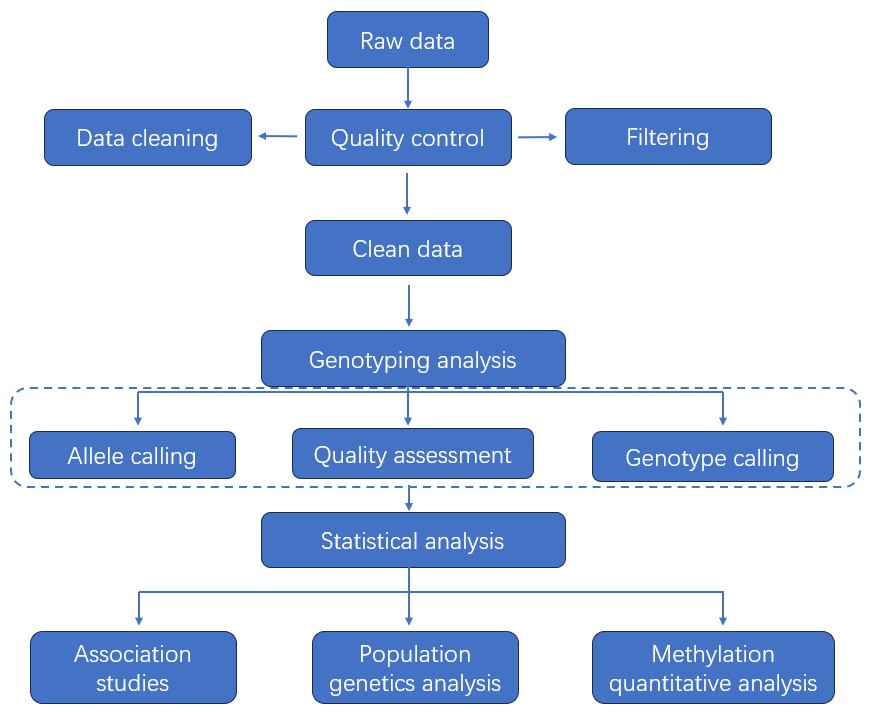
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
Our service includes customized SNP panel design, oligo manufacture and pool balancing, iPLEX chemistry running, analysis of genotyping, and delivery of genotype report in Excel format. Our workflows can be designed to accommodate both small and large assay requirements. If you have additional requirements or questions, please feel free to contact us.
References:
- Aierken K, Dong Z, Abulimiti T, et al. CDK6 3'UTR polymorphisms alter the susceptibility to cervical cancer among Uyghur females. Mol Genet Genomic Med. 2019 Mar 4:e626.
- Cai J, Cui K, Niu F, et al. Genetics of IL6 polymorphisms: Case-control study of the risk of endometrial cancer. Mol Genet Genomic Med. 2019 Mar 3:e600.
- Ji H, Lu L, Huang J, et al. IL1A polymorphisms is a risk factor for colorectal cancer in Chinese Han population: a case control study. BMC Cancer. 2019 Feb 28;19(1):181.
Demo Results
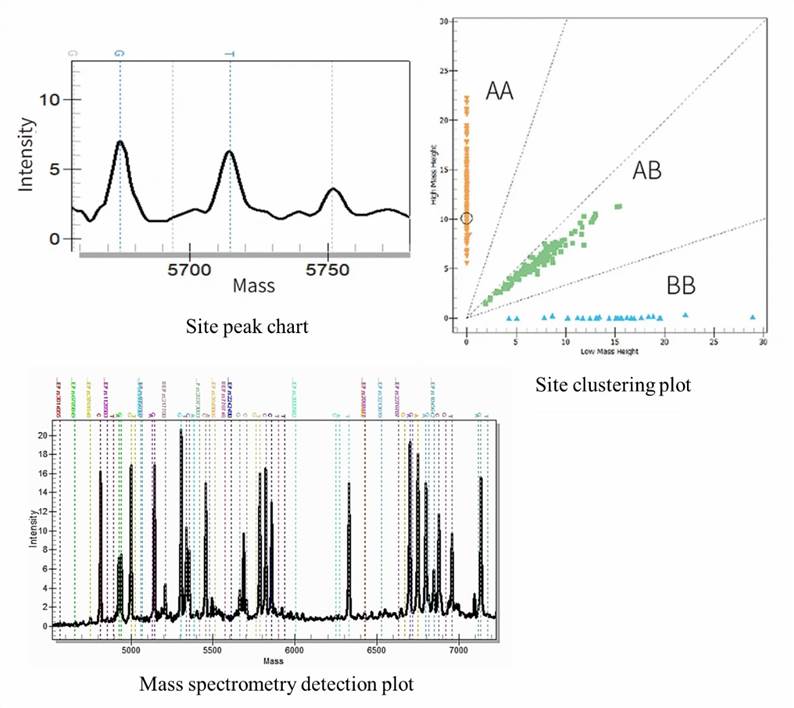
MassARRAY SNP Genotyping FAQs
1. How many samples can be processed on the MassARRAY System?
It depends on the assay design and the format of the SpectroCHIP Array being utilized. The MassARRAY System processes 2 chips per run. The chip comes in one of two formats, 96- or 384-well format. If the assay design calls for a single well, the number of samples processed in a single run ranges from 768 samples with the 384-well format chips to 192 samples with the 96-well format.
2. How accurate and reproducible are the experimental results of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF)?
Under certain experimental conditions, the accuracy and reproducibility of SNP genotyping detection results can both reach above 99%.
3. How reliable is the MassARRAY SNP Assay?
Developed by the American company Sequenom, the MassARRAY technology has found a wide array of applications in disease research, molecular diagnostics, and molecular breeding. Additionally, research utilizing this technology has been published in reputable scientific journals such as Nature, Science, and Blood.
4. What is the principle of MassARRAY?
The MassARRAY gene analysis technology is predicated upon the MALDT-TOF mass spectrometry technique. The analytical principle of this technology hinges on identifying base variations at specific SNP locations via the weight difference of individual bases. Further, it exhibits the capability to detect DNA within the approximate mass range of 4500da to 9000Da, with a mass separation resolution of 16Da.
MassARRAY SNP Genotyping Case Studies
A two-stage genome-wide association study to identify novel genetic loci associated with acute radiotherapy toxicity in nasopharyngeal carcinoma
Journal: Molecular Cancer
Impact factor: 15.302
Published: 23 August 2022
Backgrounds
Nasopharyngeal carcinoma (NPC) primarily affects southern China, with radiotherapy being a key treatment despite significant acute and late toxicities. This study conducted a two-stage GWAS on 1084 NPC patients to identify genetic variants linked to acute radiotherapy toxicity and assess their clinical predictive performance.
Methods
Sample Preparation:
- 1084 patients
- Responders
- Non-responders
Experimental Method:
- Statistical analysis
- Principal component analysis (PCA)
- Multivariate logistic regression analysis
- Linkage disequilibrium analysis
Results
The GWAS analysis identified 16 SNPs associated with five toxicities, with five SNPs validated. The study found no significant SNPs for myelosuppression, oral mucositis, and salivary gland toxicity in the validation cohort, ensuring high data quality and ethnic homogeneity among patients.
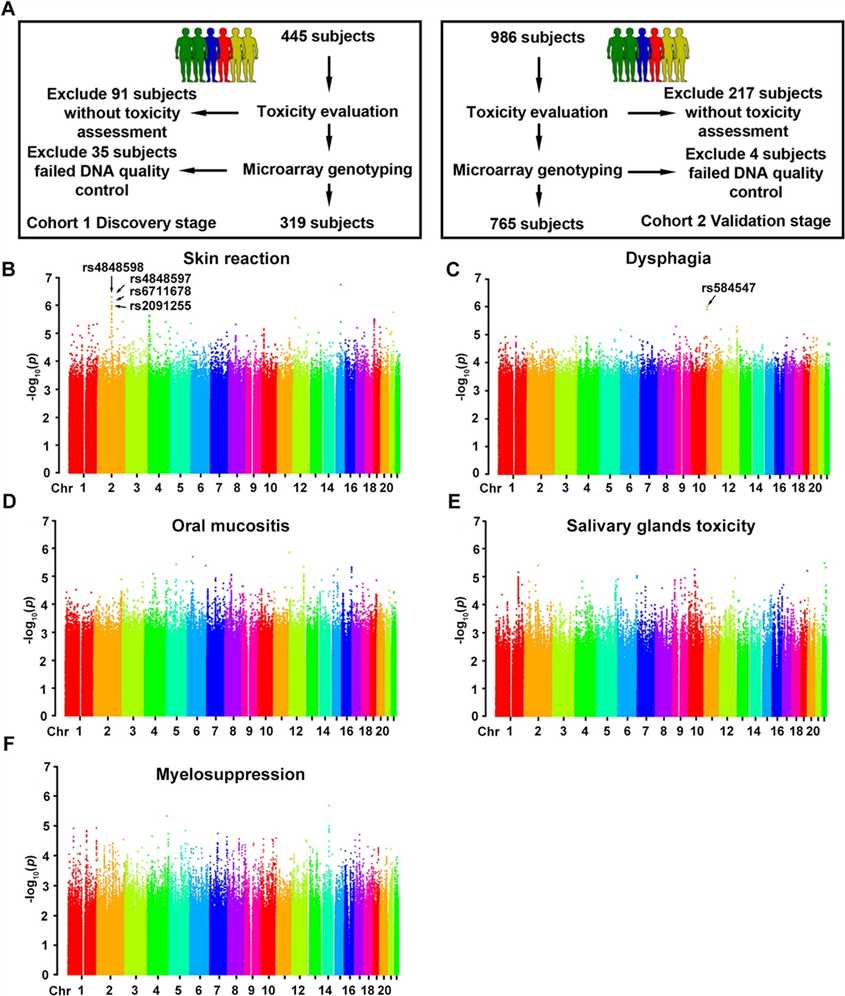 Fig 1. Genome-wide association study design and results in discovery cohort.
Fig 1. Genome-wide association study design and results in discovery cohort.
Minor allele carriers had increased risk and higher resistance to radiotherapy. Rs584547 showed a similar pattern without stratification, with higher grade 3 toxicity and resistance in A allele carriers.
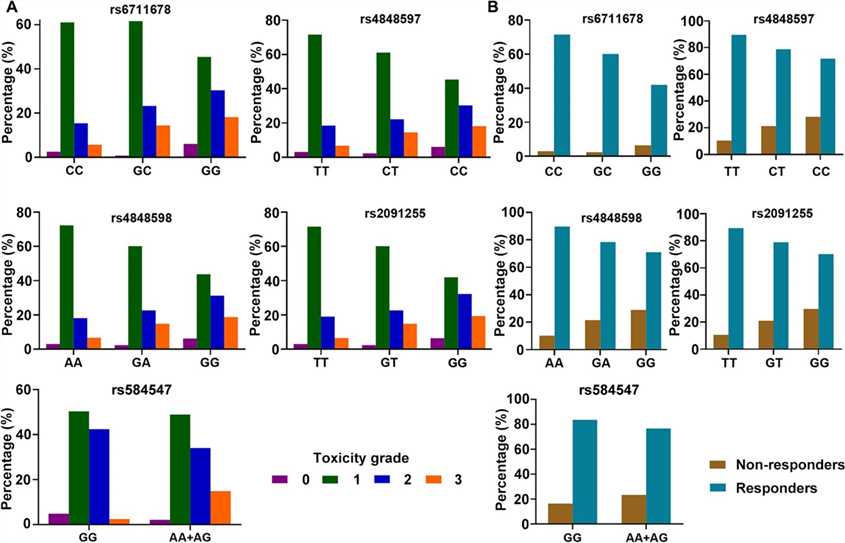 Fig 2. Patient distribution in different genotypes according to skin reaction toxicity (A) and radiotherapeutic response (B).
Fig 2. Patient distribution in different genotypes according to skin reaction toxicity (A) and radiotherapeutic response (B).
Five genetic variants were identified as potential biomarkers for predicting radiotherapy-induced skin reaction and dysphagia in NPC patients, with models combining genetic and clinical factors showing the highest accuracy.
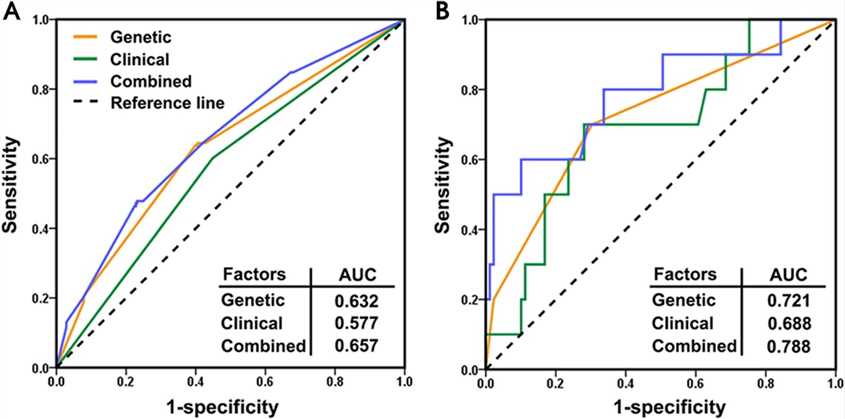 Fig 3. Receiver operating characteristic curves of three models for skin reaction (A) and dysphagia (B).
Fig 3. Receiver operating characteristic curves of three models for skin reaction (A) and dysphagia (B).
The five risk loci were mapped to genes, with four SNPs on chromosome 2q14.2 linked to LINC01101 and INHBB, while rs584547 was found to regulate SYT8 gene expression.
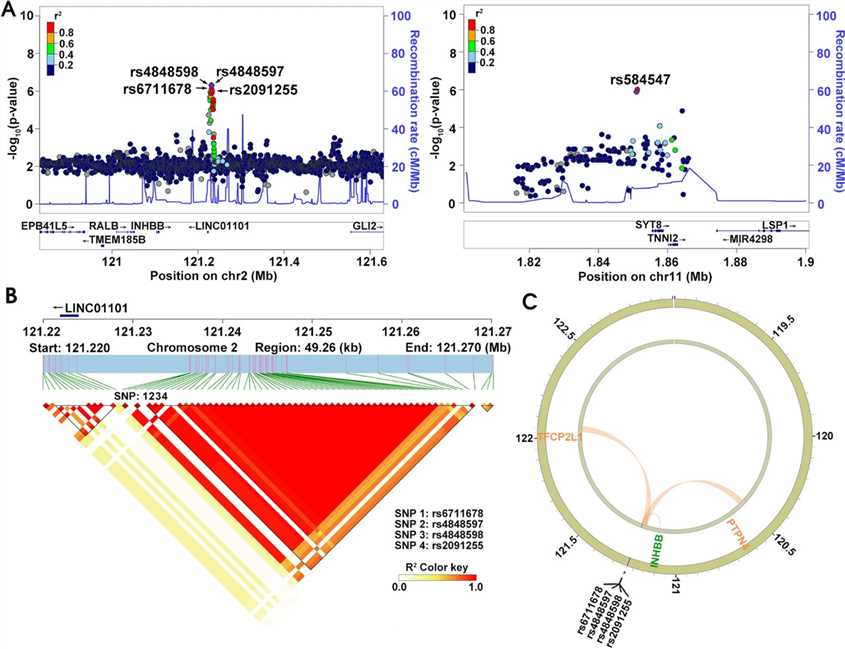 Fig 4. Gene mapping and LD analysis of rs6711678, rs4848597, rs4848598, rs2091255, and rs584547.
Fig 4. Gene mapping and LD analysis of rs6711678, rs4848597, rs4848598, rs2091255, and rs584547.
Conclusion
The authors identified five genetic variants associated with radiotherapy acute toxicities in NPC patients, which could help optimize treatment strategies to reduce these toxicities.
Reference
- Wang Y, Xiao F, Zhao Y, et al. A two-stage genome-wide association study to identify novel genetic loci associated with acute radiotherapy toxicity in nasopharyngeal carcinoma. Molecular Cancer. 2022, 21(1):169.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Investigating interactive effects of worry and the catechol-o-methyltransferase gene (COMT) on working memory performance
Journal: Cognitive, Affective, & Behavioral Neuroscience
Year: 2021
Eye color prediction using the IrisPlex system: a limited pilot study in the Iraqi population
Journal: Egyptian Journal of Forensic Sciences
Year: 2020
Embryonic origin and genetic basis of cave associated phenotypes in the isopod crustacean Asellus aquaticus
Journal: Scientific Reports
Year: 2023
Scanning indels in the 5q22.1 region and identification of the TMEM232 susceptibility gene that is associated with atopic dermatitis in the Chinese Han population
Journal: Gene
Year: 2017
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
