LncRNA Microarray Service
CD Genomics presents an extensive LncRNA (Long non-coding RNA) microarray profiling service, utilizing some of the most advanced platforms available in the field. Our methodologies ensure unparalleled accuracy and sensitivity. Through rigorous, stepwise quality control procedures, we are committed to delivering highly reliable and reproducible results.
Introduction to LncRNA Microarray
LncRNAs are a diverse and extensive class of transcribed RNA molecules, each over 200 nucleotides in length, that do not encode proteins. Their expression is intricately regulated during development and can be highly specific to certain tissues or cell types. A notable fraction of lncRNAs reside exclusively within the nucleus.
These molecules are believed to play significant regulatory roles, adding new layers of complexity to our understanding of genomic regulation. The specificity and regulatory functions of lncRNAs highlight their importance in the intricate network of gene expression.
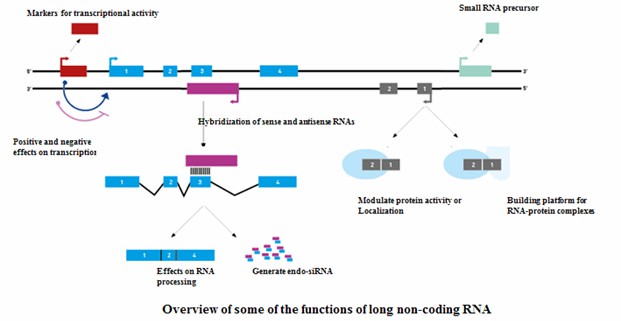
LncRNA microarray technology stands out as a sophisticated tool for analyzing lncRNA expression across diverse biological scenarios. Unlike RNA sequencing, which may falter with low-abundance lncRNAs, microarrays excel in high-throughput profiling. These arrays are engineered to detect thousands of lncRNAs alongside protein-coding mRNAs in a single experiment. By utilizing advanced probe designs and extensive lncRNA databases, microarrays enable an in-depth assessment of lncRNA expression dynamics.
CD Genomics provides a premier lncRNA microarray service that merges cutting-edge technology with rigorous quality control. The service encompasses every step from sample preparation and array hybridization to thorough data analysis, ensuring that results are both high-quality and dependable.
Advantages and Features of LncRNA Microarray
- Exceptional Sensitivity and Precision: Microarrays detect low-abundance lncRNAs with high accuracy, outperforming RNA-seq in quantifying these subtle expressions.
- Extensive Data Coverage: CD Genomics uses comprehensive and up-to-date databases like GENCODE and RefSeq, ensuring broad and current lncRNA coverage.
- Thorough Annotation and Analysis: The service includes detailed annotation and analysis, covering genomic context and subcellular localization to elucidate lncRNA functions.
- Tailored Solutions: CD Genomics offers customizable microarray solutions to suit specific research needs, providing flexible and personalized options.
- Get high quality data fast and without hassles
- A unique final report with free basic data analysis
- Data and reports delivered via secure ways
Applications of LncRNA Microarray
- Global profiling of long transcripts
- Cancer research
- Cardiovascular diseases
- Neurological disorders
LncRNA Microarray Service Workflow
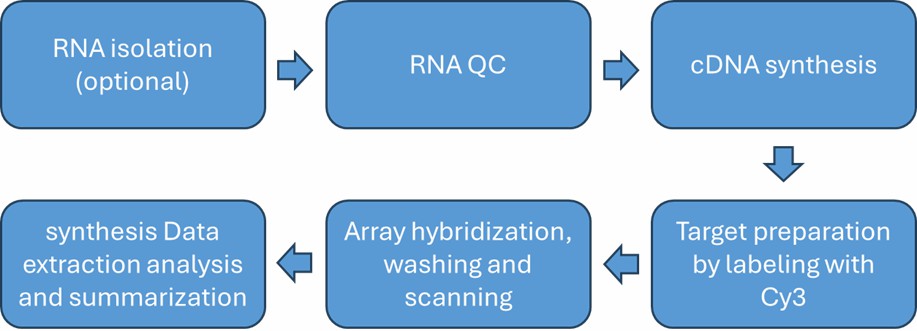
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
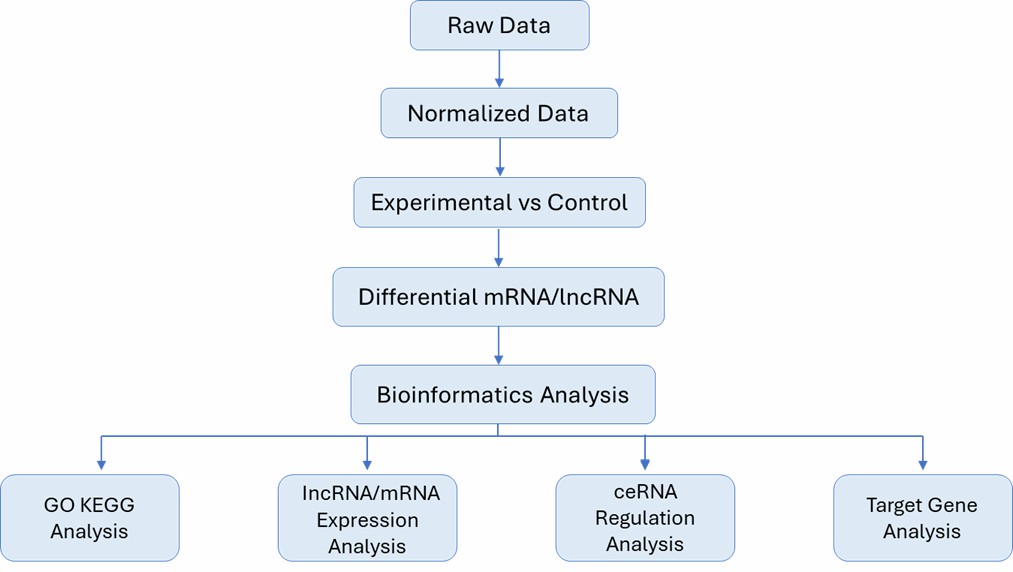
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses, including but not limited to:
|
Recommendations and Custom Service
Table1 Agilent LncRNA expression microarrays
| Microarray | Format(s) | Detected LncRNAs |
| SurePrint G3 Human Microarray | 8 х 60K | LncRNA (~17112) + genes (~22074) |
| SurePrint G3 Mouse Microarray | 8 х 60K | LncRNA (~10802) + genes (~25179) |
| Others (please click here ) | - | - |
Analysis Pipeline
Deliverables
- The raw data
- Experimental results
- Data analysis report
- Details in LncRNA Microarray for your writing (customization)
CD Genomics can also help create your custom LncRNA microarray. We are ready to help you with your custom array needs, whether it's a standard design or something more creative.
So, what you need is just to send us your samples, we can offer you the qualified final report. For details, please feel free to contact us with any questions at any time by completing a no-obligation quote request.
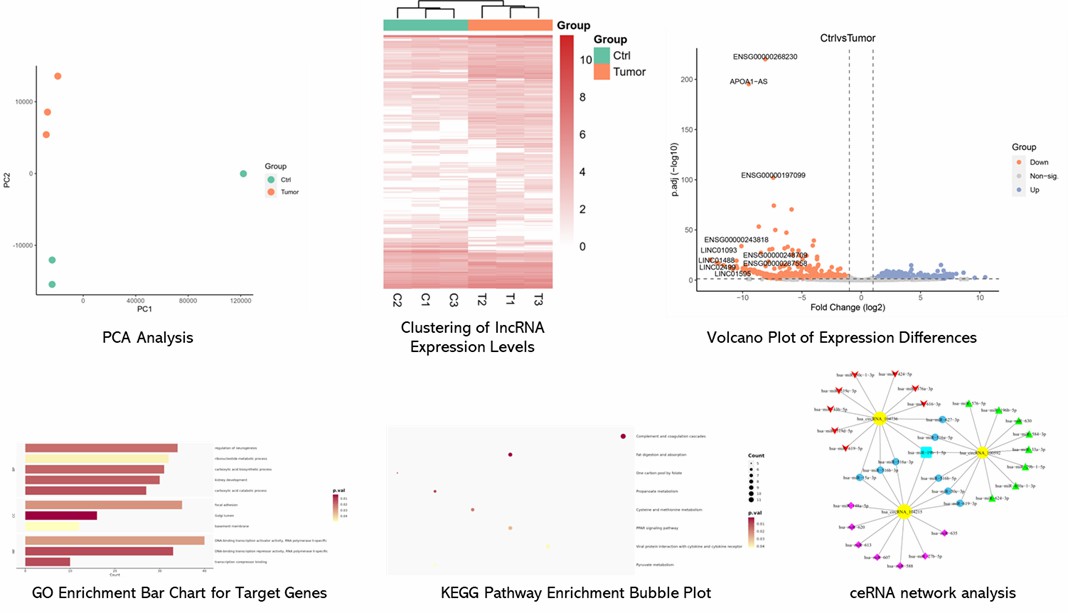
Partial results are shown below:
1. What challenges are associated with detecting lncRNAs compared to mRNAs?
The detection of long non-coding RNAs (lncRNAs) presents distinct challenges relative to messenger RNAs (mRNAs), primarily due to their inherently lower expression levels and pronounced tissue specificity. The unique attributes of lncRNAs, such as their low abundance and extensive isoform diversity, introduce additional complexity to their analysis. To effectively surmount these obstacles, our microarrays have been meticulously designed to ensure the precise detection and quantification of lncRNAs, despite their intricate nature.
2. How does the sensitivity of lncRNA microarrays compare to RNA sequencing?
In terms of sensitivity, lncRNA microarrays exhibit superior performance in detecting low-abundance lncRNAs when compared to RNA sequencing. RNA sequencing can encounter accuracy issues, particularly arising from high error rates and the inherently low expression levels of these transcripts. In contrast, our microarrays provide reliable measurements with minimal susceptibility to variations caused by low transcript abundance, thereby enhancing the overall sensitivity of detection.
3. Can lncRNA microarrays detect different transcript isoforms?
Absolutely, our lncRNA microarrays are adept at differentiating and detecting various isoforms. Unlike mRNAs, lncRNAs are known to generate multiple isoforms, each with uniquely distinct biological roles. Our advanced microarray design ensures the accurate identification and quantification of these diverse isoforms, thereby offering a comprehensive understanding of lncRNA functionality.
4. How does the accuracy of lncRNA quantification in microarrays compare to sequencing methods?
When it comes to the quantification of low-abundance lncRNAs, microarray technology offers greater accuracy compared to sequencing methods, which are often hampered by higher error rates and coverage limitations. Our microarrays are rigorously engineered to mitigate these issues, resulting in the provision of more reliable and extensive data that significantly enhances the precision of lncRNA quantification.
5. What custom solutions are available for lncRNA microarray analysis?
We offer tailored lncRNA microarray solutions that cater to specific research needs. Whether your requirements involve standard arrays or innovative designs, our customizable options ensure that our services are precisely aligned with your research objectives. This flexibility facilitates the acquisition of targeted and comprehensive genomic insights, thereby advancing your scientific endeavors.
Integrated Analysis of lncRNA and mRNA Expression Profiles Indicates Age-Related Changes in Meniscus
Journal: Frontiers in cell and developmental biology
Impact Factor: 5.206
Published: 10 March 2022
Background
The meniscus, crucial for knee function, deteriorates with age, heightening osteoarthritis risk due to changes like collagen thickening. Long non-coding RNAs (lncRNAs) are important in tissue aging and may contribute to meniscus degeneration. To explore this, the study analyzed lncRNA and mRNA expression profiles in young and aging menisci using microarray analysis, followed by bioinformatics to identify key molecules and pathways involved in age-related meniscus changes.
Materials & Methods
Sample Preparation
- Human meniscus sample
- Histology staining
- RNA extraction
Method
- Microarray Hybridization
- Arraystar, 8 × 60K
- Functional enrichment analysis of differentially expressed mRNAs
- Protein and protein interaction analysis
- LncRNAs-mRNAs co-expressed analysis
- Statistical analysis
Results
The study used Arraystar Human lncRNA and mRNA Array to compare meniscus tissues from young and aging groups (n = 4/group). It identified 1608 differentially expressed lncRNAs and 1809 differentially expressed mRNAs. Expression patterns were visualized with volcano plots and hierarchical clustering.
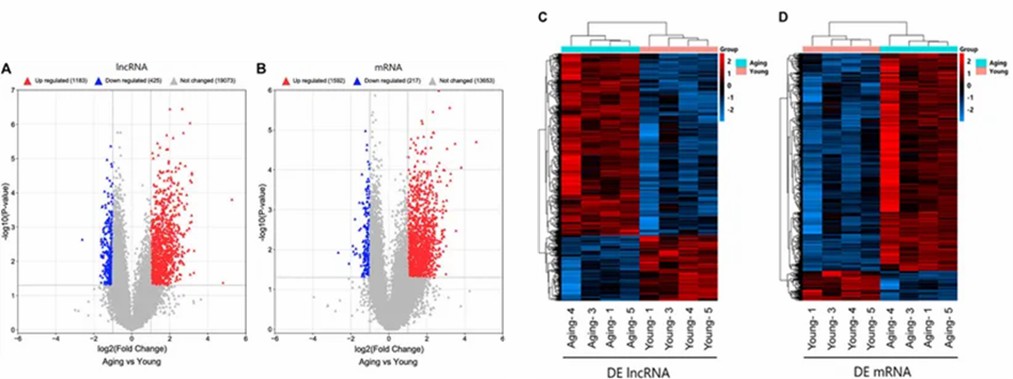 FIGURE 1. Identifification of DE lncRNAs and mRNAs.
FIGURE 1. Identifification of DE lncRNAs and mRNAs.
GO and pathway analyses of 1809 differentially expressed mRNAs highlighted key pathways involved in cartilage damage, including TGF-beta, Wnt, and PI3K-Akt. The study identified 18 mRNAs significantly altered, with notable downregulation of FGF6 and PPP2R5C.
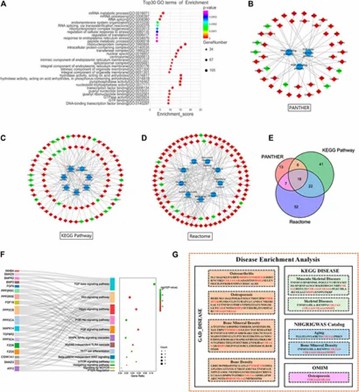 FIGURE 2. Functional Enrichment Analysis of DE mRNAs.
FIGURE 2. Functional Enrichment Analysis of DE mRNAs.
A ceRNA network analysis using 19 DE mRNAs and 46 DE lncRNAs identified 98 nodes and 229 edges. The network, constructed with Cytoscape, highlighted lncRNA POC1B-AS1, which was further examined for its miRNA interactions and target genes in a detailed subnetwork.
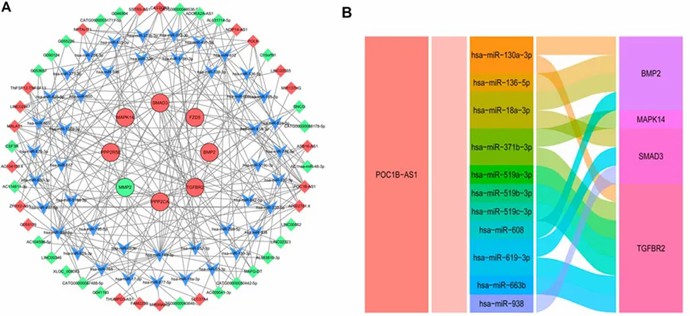 FIGURE 3. CeRNA Analysis.
FIGURE 3. CeRNA Analysis.
Conclusion
This study found significant changes in lncRNA and mRNA expression in the aging meniscus, including decreased MMP-2 and COL1A2 levels. Key pathways like Hippo and TGF-beta are involved in meniscal degeneration. Abnormal lncRNA expressions suggest new therapeutic targets for treating meniscal degeneration.
Reference
- Ai LY, Du MZ, Chen YR, et al. Integrated analysis of lncRNA and mRNA expression profiles indicates age-related changes in meniscus. Frontiers in cell and developmental biology. 2022, 10:844555.
Here are some publications that have been successfully published using our services or other related services:
Use of biostimulants for water stress mitigation in two durum wheat (Triticum durum Desf.) genotypes with different drought tolerance
Journal: Plant Stress
Year: 2024
The Restriction-Modification Systems of Clostridium carboxidivorans P7
Journal: Microorganisms
Year: 2023
In the land of the blind: Exceptional subterranean speciation of cryptic troglobitic spiders of the genus Tegenaria (Araneae: Agelenidae) in Israel
Journal: Molecular Phylogenetics and Evolution
Year: 2023
Genetic Modifiers of Oral Nicotine Consumption in Chrna5 Null Mutant Mice
Journal: Front. Psychiatry
Year: 2021
A high-density genetic linkage map and QTL identification for growth traits in dusky kob (Argyrosomus japonicus)
Journal: Aquaculture
Year: 2024
Genomic and chemical evidence for local adaptation in resistance to different herbivores in Datura stramonium
Journal: Evolution
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
