DNA Methylation Assay (Illumina 935K) Service
Devolopment of DNA Methylation Assay
DNA methylation plays a crucial role in regulating gene expression, with its state changes intricately intertwined with aging, development, and pathological processes associated with diseases. To assess DNA methylation, there exists a pressing need for an approach offering high-throughput, single-base resolution metamorphic detection, applicable across various species and sample types, including formalin-fixed, paraffin-embedded (FFPE) samples and circulating free DNA (cfDNA) in blood. In this regard, methylation microarrays, being convenient and cost-effective preliminary screening tools, provide significant advantages.
Over the past decade, Illumina Inc. has leveraged its BeadArray™ technology to furnish researchers with powerful microarray tools designed to conduct quantitative analyses of whole-genome DNA methylation. In this aspect, the Infinium Human Methylation 450 (450K) and Methylation EPIC v1.0 BeadChips (850k) have been extensively used to facilitate data collection in Epigenome-Wide Association Studies (EWAS). These methylation microarrays have propelled the discovery and application of methylation-based biomarkers in cancer research, genetic diseases, aging, molecular epidemiology, and several other research fields.
Within this context, Illumina has introduced an upgraded version of the methylation chip – the Infinium MethylationEPIC v2.0 BeadChip (935k). Retaining the reliable chemical foundation of the original Infinium whilst updating and optimizing numerous methylation sites, this newest chip strives to stimulate more biological discoveries in the new epoch of epigenetic research.
The transition from the 450k to the 850k, and now to the current 935k methylation arrays has seen a continual increase in the detectable methylation sites. So, what novel developments are evident in the 935k compared to its predecessors?
The EPIC v2.0-935k array can detect the methylation status of approximately 935,000 CpG sites across the human genome. It builds on the basis of the EPIC v1.0-850k by eliminating sub-performing probes, adding 186,000 CpG targets to enhancers and super-enhancers, and including more CTFC binding sites, CNV detection regions, CpG islands with insufficient coverage in EPIC v1.0, and common cancer-driving mutations. Additionally, the updated array aligns with the HG38 genome version and employs the v41 version of the GenCode database, making it more suitable for EWAS research.
Human DNA Methylation Assay 935K Service at CD Genomics
CD Genomics has experimentally tested the new product using low-quality and easily-degradable FFPE samples. By investigating the detection rate and quality control data from various experimental stages, we can confirm the applicability of this new product and the reliability of CD Genomics' detection capabilities.
Advantages of DNA Methylation Assay 935K Service
Whole-Genome Analysis
Following a comprehensive evaluation of EPIC v1.0 by expert analysts, considerable enhancements have been made to the initial probe features. After eliminating lower-performing probes, cutting-edge research proceedings were incorporated, considerably augmenting EPIC's capabilities in the area of epigenetic research. EPIC v2.0 introduces an additional 186,000 CpG target enhancers and super enhancers, and broadens the CTCF binding sites, CNV detection zones, and supplements the underrepresented CpG island data found in EPIC v1.0. Furthermore, version 2.0 integrates information on driver mutations related to common cancers.
Simultaneously, the upgraded v2.0 Beadchip is capable of analyzing chromatin open areas identified through ATAC-Seq and ChIP-seq experiments, providing scientific researchers with more abundant and precise data support. The features include:
- Different methylation positions discovered in various cancers’ tumour and normal samples.
- Enhancers and super enhancers identified in cancer and cell line samples through ChIP-Seq.
- Different accessible chromatin regions recognized in primary human cancers using ATAC-Seq.
- Expansion of CpG island coverage.
- Enhancement of exon coverage to enable more accurate CNV detection.
- Mutations in drive genes associated with common cancers.
Trustworthy technology
Illumina Methylation BeadChip has achieved breakthrough research results in the field of epigenetics over the past decade. At present, most published epigenome-wide association study (EWAS) depend on MethylationEPIC or its predecessor, HumanMethylation450. Consequently, researchers can take full advantage of abundant methylation chip data in public databases such as GEO and TCGA, further accelerating research progress.
Compatible with multiple sample types, including FFPE
After meticulous design and validation, the Infinium methylation assay demonstrates superior reliability, as evidenced by the high-quality data obtained from FFPE tissue samples. Given the pivotal role of FFPE samples in cancer research, their compatibility is crucial for advancing research based on large tumor biosample libraries. Notably, the Infinium assay is not only applicable to FFPE samples, but also accommodates a variety of sample types including fresh/frozen tissues, whole blood, oral cells, cfDNA, and saliva, ensuring its widespread application across diverse research scenarios.
Data Analysis
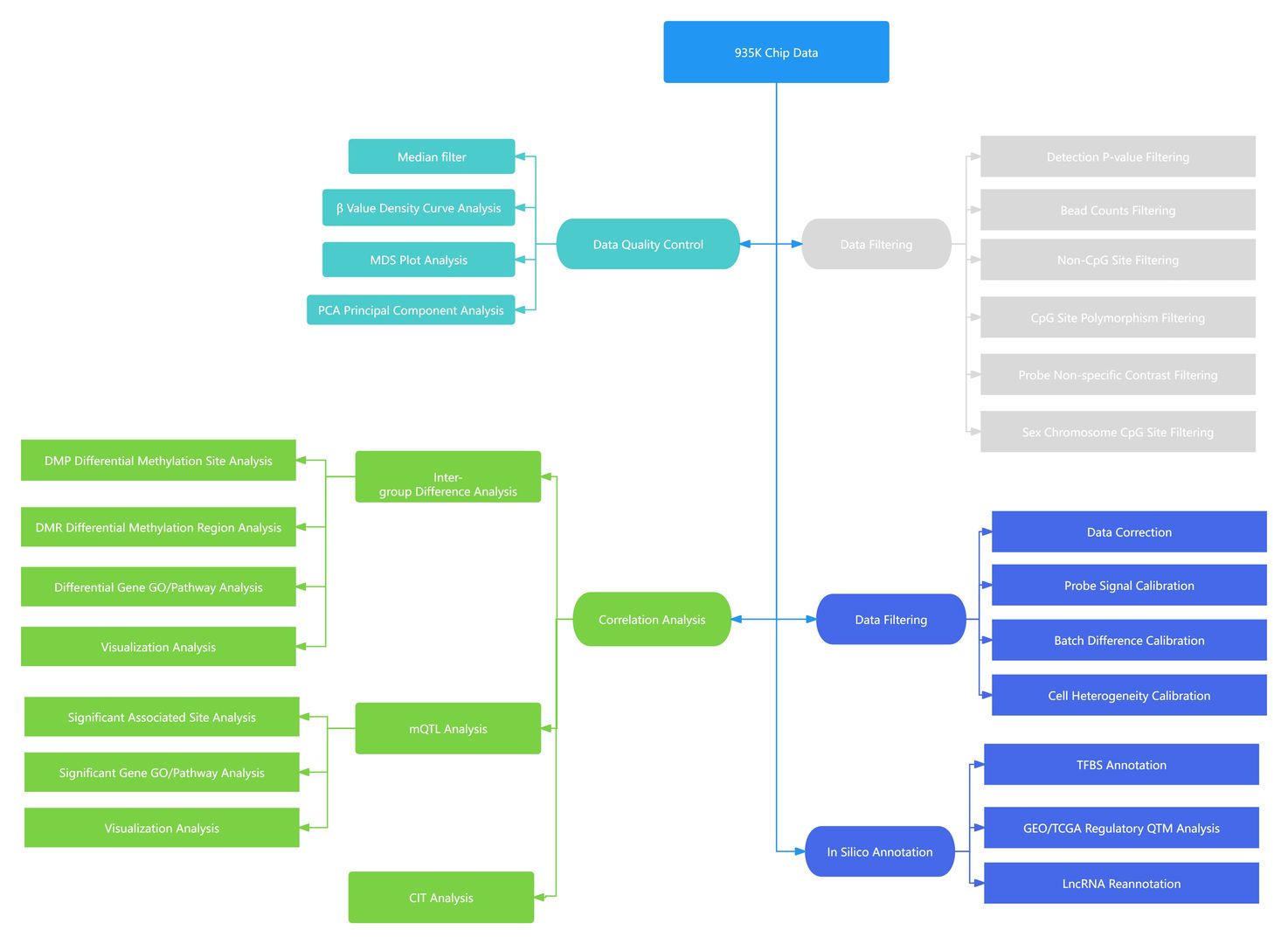
Sample Requirement
| Sample Type | Sample Requirements |
|---|---|
| Human, Animal Genomic DNA | Provide ≥2ug; remove RNA and protein residues; concentration ≥20ng/ul, based on Qubit quantification |
| Fresh Frozen Tissue | Provide not less than 10-30mg tissue; store at -20°C or -80°C freezer |
| Cultured Cells | Provide not less than 2x10^6 cells; collect adherent or suspended cells, wash once with PBS, centrifuge at 600g for 5 minutes, remove supernatant PBS after centrifugation, retain cell pellet |
| Whole Blood Sample | Provide 0.5-2ml peripheral blood or buffy coat; place in 1.5/2.0ml centrifuge tube; avoid tubes containing anticoagulants that may interfere with analysis |
| Paraffin-embedded FFPE | Provide ≥1ug; fragment main band larger than 500bp; concentration ≥20ng/ul; based on Qubit quantification |
| Provide not less than 15 slides, tissue area not less than 1cm², 4-10μM thick; tumor cell proportion not less than 30%; place slides in 1.5/2.0 ml centrifuge tube | |
| Plasma/Serum/cfDNA | Provide >15ng cfDNA, no large fragment genome, mononucleosome cfDNA proportion not less than 50% |
| Provide 1-4ml plasma/serum |
DNA Methylation Assay (Illumina 935K) Service Case Studies
Methylation profiling identifies two subclasses of squamous cell carcinoma related to distinct cells of origin.
Journal: NATURE COMMUNICATIONS (IF=16)
Technology: Illumina DNA methylation chip and RNA-seq.
Research Background: Squamous cell carcinoma (cSCC) is a malignant tumor that occurs in epidermal or adnexal cells. It is a prevalent type of skin cancer, usually caused by the progression of precancerous lesions induced by ultraviolet radiation, known as actinic keratosis (AK). The majority of epigenetic changes associated with cSCC are primarily composed of a moderate number of cancer-related genes, silenced by high CpG island promoter methylation. However, only partial understanding exists towards the etiology of AK and its crucial transition process into cSCC.
Research Findings: This study explores DNA methylation changes during the progression from healthy epidermal cells to AK and finally to Cutaneous SCC. It provides the most comprehensive epigenomic analysis of cSCC development to date. The findings highlight the existence of two distinct subclasses of cSCC, reflecting differentiation stages of diverse cell origins.
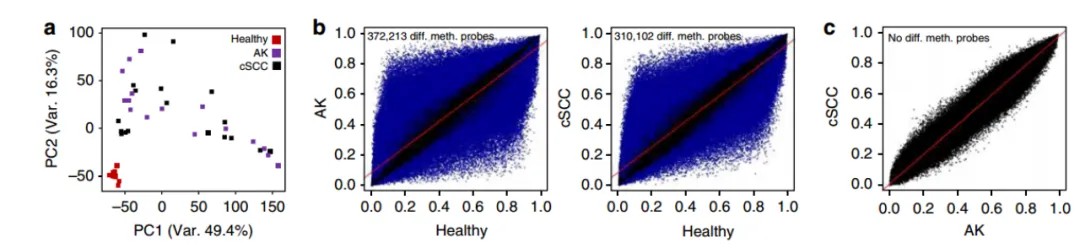
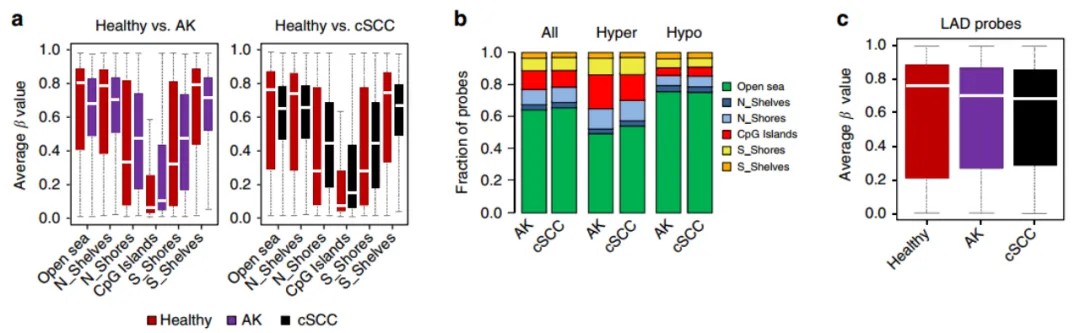
Reference:
- Rodríguez-Paredes, M., Bormann, F., Raddatz, G. et al. Methylation profiling identifies two subclasses of squamous cell carcinoma related to distinct cells of origin. Nat Commun 9, 577 (2018). https://doi.org/10.1038/s41467-018-03025-1
DNA Methylation Assay (Illumina 935K) Service FAQs
Q1: What sites from 850K chip are preserved in the design of 935K chip?
A2: In the upgrade to the methylation chip, the following site information have been retained: 1) CpG islands 2) Non-CpG methylation sites in human stem cells (CHH sites) 3) ENCODE open chromatin and enhancers 4) FANTOM5 enhancers 5) Deoxynucleotidyl transferase hypersensitive sites 6) miRNA promoter regions 7) It covers more than 85% of sites in the Infinium Methylation 450 BeadChip.
Q2: What additional loci does the 935K chip incorporate on the basis of the 850K chip?
A2: The following loci have principally been added: 1) Differential methylation sites identified in tumor samples and normal samples across multiple cancers. 2) Enhancers and super-enhancers identified in cancer and cell line samples through ChIP-Seq. 3) Differentially accessible chromatin regions identified in primary human cancers with ATAC-Seq. 4) Expanded coverage of CpG islands. 5) Enhanced exome coverage that allows for accurate detection of CNVs. 6) Common cancer driver mutations.
Q3. Can the data from the 935K chip be analyzed together with the data from the 850K chip?
A3: Yes, overlapping sites from the 850K chip and the 935K chip as listed in question 1 can be analyzed together.
Q4. Does the type of samples the 935K chip can process differ from those that the 850K chip can handle?
A4: No, types of samples capable of being processed by the 850K chip can equally be processed by the 935K chip.
Q5. What is the requirement for the number of samples in a single experiment?
A5: ZKPR is currently one of the largest methylation chip platforms in the country, and it does not impose special restrictions on the number of samples.
Q6. What are the application fields of methylation chips?
A6: Methylation chips are primarily used in the biomedical domain, such as: various cancer research (lung cancer, esophageal cancer, ovarian cancer, etc.); complex disease research (diabetes, hypertension, autoimmune diseases, rare diseases, psychiatric diseases, etc.); developmental research (embryonic development, neural differentiation, organ differentiation, etc.); environmental interactions (smoking, alcohol consumption, industrial exposure, pollution, etc.); aging (methylation age); cfDNA tumor screening (various biomarker screenings), and more.


 Sample Submission Guidelines
Sample Submission Guidelines
