What Is Plant and Animal Whole Genome de novo Sequencing?
Plant and animal whole genome de novo sequencing refers to assembling a complete genome without relying on any existing reference sequence. This approach is essential when working with species that lack a reference genome, have poorly assembled genomes, or exhibit complex genomic features like high heterozygosity, polyploidy, or extensive repetitive regions.
Instead of aligning sequencing reads to a known genome, de novo assembly reconstructs the genome from scratch—like solving a massive jigsaw puzzle using only the sequence fragments generated by high-throughput sequencing platforms. The result is a high-resolution genetic blueprint that can be used for functional annotation, comparative analysis, molecular breeding, and evolutionary studies.
At CD Genomics, we provide end-to-end de novo sequencing services for a wide range of plant and animal species. By integrating Illumina short reads, PacBio HiFi long reads, Nanopore ultra-long reads, and Hi-C chromatin interaction data, we deliver chromosome-level genome assemblies ready for downstream research and publication.
When Should You Use De Novo Genome Sequencing?
De novo genome sequencing is the preferred strategy when no high-quality reference genome exists—or when existing references cannot meet your research objectives. Here are the most common use cases:
✅ No Reference Genome Available
For newly discovered or under-studied species, de novo sequencing enables researchers to generate a complete reference from scratch.
✅ Incomplete or Fragmented Reference
Many publicly available genomes are outdated, poorly assembled, or fragmented at the scaffold level. De novo assembly delivers chromosome-level continuity for high-resolution research.
✅ Complex Genomes: Polyploidy, Heterozygosity, Repeats
Plant and animal genomes often contain high levels of duplication, structural variation, or repetitive elements. De novo approaches using long-read sequencing and Hi-C mapping overcome these challenges.
✅ Pan-Genome Construction
When a single reference genome cannot capture the genetic diversity of a species, building a pan-genome via de novo assembly of multiple individuals reveals population-specific variation.
✅ Trait Discovery and Molecular Breeding
High-quality assemblies provide the foundation for GWAS, QTL mapping, and genome editing—especially in agricultural, aquaculture, and livestock research.
Pro Tip:
De novo sequencing is not only for novel species. It is often the best way to upgrade a low-contiguity genome to publication-ready quality, especially when combined with HiFi and Hi-C data.
Technology Strategy Overview: Platform Comparison for Genome Assembly
| Platform | Role in Assembly | Typical Coverage | Strengths | Recommended For |
|---|---|---|---|---|
| Illumina / DNBSEQ™ | Genome survey, error correction | 30–50× | High accuracy, low cost, essential for k-mer profiling | Initial genome complexity analysis |
| PacBio HiFi | Contig-level de novo assembly | 30–60× | Ultra-high accuracy (Q20+), excellent for repeat-rich or polyploid genomes | Plant/animal genomes with high heterozygosity |
| Oxford Nanopore (ONT) | Gap closure, ultra-long read assembly | 50–100× | Ultra-long reads (>100 kb), ideal for telomere-to-telomere (T2T) assemblies | Genomes requiring complete or near-complete continuity |
| Hi-C | Chromosome-scale scaffolding | 100–150× | Builds chromosome pseudomolecules, corrects misassemblies | Final chromosome anchoring and QC |
| 10x Genomics Linked-Reads | Repeat resolution, phasing (optional) | ~60× | Phases heterozygous loci, supports haplotype separation | Diploid or highly heterozygous species |
| BioNano Optical Mapping | Large structural variation detection (optional) | NA | Detects SVs, scaffolds complex assemblies | Very large or structurally complex genomes |
 Hybrid Strategy Insight:
Hybrid Strategy Insight:
Most successful assemblies combine short reads + long reads + Hi-C. We tailor the platform mix based on genome size, ploidy, and your research goals.
De Novo Genome Sequencing Service Workflow: From Sample to Chromosome-Scale Assembly
Sample Quality Control
- Integrity assessment via PFGE or Femto Pulse
- Purity checks (OD ratios, Qubit, and RNA contamination removal)
Genome Survey (Illumina)
- Short-read sequencing (~100X coverage)
- K-mer analysis for genome size, repeat content, heterozygosity
- Guides downstream long-read and Hi-C strategy
Long-Read Sequencing (PacBio HiFi or Oxford Nanopore)
- High-contiguity de novo assembly of primary contigs
- Platforms selected based on target genome properties
- 30–100X coverage depending on platform
Scaffolding and Chromosome Anchoring (Hi-C Sequencing)
- Captures long-range chromatin interactions
- Anchors contigs to pseudochromosomes
- Enables chromosome-scale genome assembly
Polishing and Error Correction
- Short-read polishing for SNP/indel correction
- Gap filling and repeat resolution
- BUSCO and alignment-based quality checks
Genome Annotation (Optional Add-On)
- Gene structure prediction (ab initio and evidence-based)
- Repeat region masking
- Functional annotation (GO, KEGG, Pfam)

Bioinformatics Analysis
Our genome informatics pipeline integrates high-throughput assembly, annotation, and comparative analysis—customized for both plant and animal species. Whether you're working with a diploid, polyploid, or highly repetitive genome, we provide scalable and accurate solutions to decode complexity.


Workflow

Sample Requirements & Quality Standards
| Sample Type | Required Amount | Purity Criteria | Special Notes |
|---|---|---|---|
| Fresh or frozen animal tissue | ≥ 1.5 μg gDNA (≥50 kb average length) | OD260/280: 1.8–2.0; OD260/230: ≥2.0 | Avoid blood-contaminated samples; no freeze-thaw cycles |
| Plant leaves or stems | ≥ 2 μg gDNA (≥50 kb average length) | Same as above | Avoid polysaccharide, polyphenol contamination; prefer young, tender tissues |
| Cultured cells (e.g., fish, insects) | ≥ 1.5 μg gDNA | Same as above | For insects, remove chitin exoskeleton before extraction |
| Hi-C crosslinked tissue | ≥ 1 g fresh tissue or ~5 million cells | OD not applicable (crosslinked) | Crosslinking and fixation must follow our Hi-C prep protocol |
General QC Criteria:
- High molecular weight DNA: >50 kb preferred for long-read platforms (PacBio HiFi, Oxford Nanopore)
- No RNA, protein, or secondary metabolite contamination
- Concentration: ≥50 ng/μL (Qubit); Integrity: Confirmed by pulsed-field gel or Femto Pulse
Need help with DNA extraction?
CD Genomics provides end-to-end extraction services tailored to plant and animal genomes, using magnetic bead purification to minimize shearing and contaminants. Contact us to learn more.
Deliverables
CD Genomics provides comprehensive and well-organized deliverables for every plant or animal whole genome de novo sequencing project. Our data packages are tailored for seamless downstream analysis and publication readiness.
✅ Standard Deliverables
| File Type / Content | Description |
|---|---|
| Raw Sequencing Data | FASTQ files from PacBio HiFi, Nanopore, Illumina, and/or Hi-C platforms |
| Assembly Results | Genome contigs and scaffolds in FASTA format |
| Assembly Metrics Report | Summary of genome size, N50, GC content, completeness (BUSCO, etc.) |
| Genome Annotation (Optional) | GFF3/GTF files, functional annotation tables, gene structure visualization |
| Hi-C Interaction Map | Contact matrices and assembly scaffolding plots (if Hi-C is included) |
| Circos & Synteny Plots | Visual summaries of genome architecture and comparative analysis |
| Bioinformatics Summary Report | Detailed methods, software versions, and pipeline descriptions |
✅ Optional Add-ons (Project Upgrades)
For projects requiring advanced data analysis or tailored outputs, CD Genomics offers the following upgrade options:
| Upgrade Option | Description |
|---|---|
| Chromosome-Level Assembly | Achieved via Hi-C or BioNano scaffolding, delivering chromosome-scale pseudomolecules |
| Functional Genome Annotation | Includes gene prediction, GO/KEGG enrichment, repeat elements, and TE annotations |
| Comparative Genomics Package | Includes whole-genome synteny, ortholog clustering, and evolutionary distance estimation |
| Pan-Genome Construction | Multi-sample assembly integration, structural variant detection, and shared/unique gene sets |
| Epigenome Integration | Add-on for methylation or histone modification maps (requires compatible sample prep) |
| GWAS-Ready Data Formatting | Includes SNP/INDEL calling, VCF formatting, and population structure files for GWAS pipelines |
Featured Project Snapshot
| Species Type | Genome Size | Contig Count | Contig N50 | Hi-C Anchoring Rate |
|---|---|---|---|---|
| Plant A | 1.02 Gb | 626 | 7.15 Mb | 95.4% |
| Plant B | 793.46 Mb | 347 | 34.19 Mb | 96.1% |
| Aquatic Animal A | 979.98 Mb | 513 | 5.36 Mb | 97.89% |
| Aquatic Animal B | 827.62 Mb | 170 | 9.88 Mb | 99.51% |
| Mammal | 3.3 Gb | 2,658 | 79.41 Mb | 98.58% |
| Insect | 979.98 Mb | 513 | 5.37 Mb | 97.89% |
These high-contiguity genomes demonstrate CD Genomics' robust assembly pipeline across diverse species—from complex plant genomes to chromosome-level assemblies in mammals and aquatic organisms.
Demo Results
Partial results are shown below:

Distribution of base quality.

Distribution of base content.

Shared SNP number between samples.

SNP mutation type distribution.
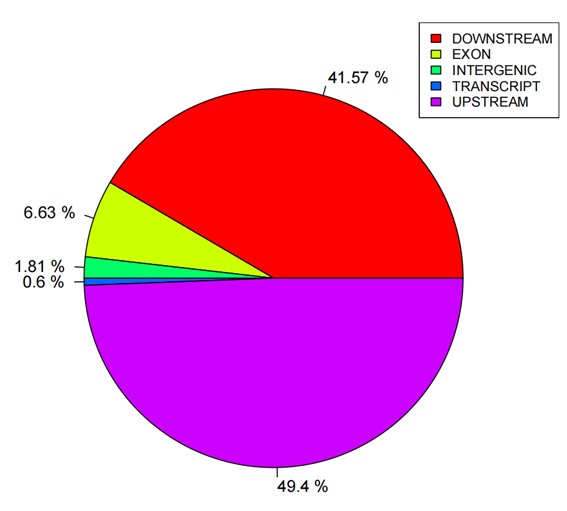
Statistics pie of SNP annotations.

Shared InDel number between samples.

InDel length distribution in both the whole genome scale and CDS regions.

Statistics pie of InDel annotations.
Plant/Animal Whole Genome de novo Seq FAQs
What is Plant or Animal Whole Genome de novo Sequencing?
It's a reference-free approach to reconstructing a species' entire genome from scratch. This method is essential for species lacking a reliable reference genome or those with complex structural variations.
When should I choose de novo genome sequencing over resequencing?
Answer:
Choose de novo sequencing when:
- No high-quality reference genome exists.
- Your species has significant genomic diversity or complexity.
- You aim to build a pan-genome or improve current reference quality.
What sequencing platforms are used in your service?
We use a hybrid strategy that combines:
- Illumina (for k-mer-based survey and polishing)
- PacBio HiFi / Oxford Nanopore (for long-read contig generation)
- Hi-C (for chromosome-level scaffolding)
This layered approach maximizes assembly continuity and accuracy.
What sample quality is required?
Typical requirements include:
- High-molecular-weight gDNA
- OD260/280 = 1.8–2.0
- OD260/230 ≥ 2.0
- ≥10–15 μg total DNA depending on platform
We provide detailed submission guidelines upon inquiry.
What deliverables will I receive?
Deliverables include:
- High-quality assembled genome (FASTA)
- Assembly metrics and quality report
- Gene prediction and functional annotation files
- Visual summaries (e.g., genome circle maps, synteny plots)
Do you offer downstream analysis?
Yes. CD Genomics provides advanced bioinformatics options including:
- Ortholog clustering
- Phylogenetic reconstruction
- Gene family expansion analysis
- Genome synteny and collinearity assessment
Plant/Animal Whole Genome de novo Seq Case Studies
Customer Publication Highlight
Case Study: Deciphering m6A Methylation Mechanisms in Arabidopsis Using Whole Genome de novo Sequencing
Journal: New Phytologist
Impact Factor: 8.3
Published: 2017
DOI: 10.1111/nph.14586
Background
As a model organism for plant genetics, Arabidopsis thaliana has been instrumental in uncovering epigenetic regulatory mechanisms. Among these, N6-methyladenosine (m6A) modification of mRNA plays a pivotal role in plant growth, development, and stress responses. However, the molecular components driving this modification—and their functional conservation in higher plants—remain incompletely understood.
This study aimed to identify the genetic factors essential for m6A RNA methylation in Arabidopsis by integrating whole genome de novo sequencing with targeted functional genomics. A central focus was placed on understanding the role of HAKAI, a conserved E3 ubiquitin ligase, within the methylation machinery.
Materials & Methods
Genome Analysis and Mutant Screening:
- Arabidopsis thaliana mutants with suspected m6A methylation defects were selected from T-DNA insertion libraries.
- Genomic DNA was extracted and sequenced de novo using Illumina short-read and ONT long-read platforms, achieving high contiguity and coverage.
- Gene disruption events were mapped and validated.
m6A Profiling:
- Total RNA was isolated from mutant and wild-type lines.
- m6A quantification was performed using LC-MS/MS and immunoprecipitation-based m6A-seq.
Functional Validation:
- Complementation assays were used to verify gene function.
- RNA-seq was applied to assess transcriptomic consequences of HAKAI loss.
Results
The whole genome de novo sequencing enabled accurate identification of T-DNA insertions disrupting HAKAI, a gene encoding a RING-domain E3 ubiquitin ligase. Functional loss of HAKAI significantly reduced global m6A methylation levels, comparable to mutants of known m6A writers such as MTA and FIP37.
Key Findings:
- Loss of HAKAI led to defects in apical dominance, flowering time, and embryo viability, phenocopying other core m6A component mutants.
- Transcriptome analysis revealed dysregulation in key developmental and hormone signaling pathways.
- Complementation of the HAKAI gene restored both m6A methylation levels and normal development.
Conclusion
This study demonstrated that HAKAI is a critical component of the m6A methylation complex in plants, acting alongside canonical methyltransferases. The use of whole genome de novo sequencing allowed precise mapping of gene disruptions and was essential for validating functional hypotheses in genetically complex backgrounds.
The case highlights how plant whole genome de novo sequencing, paired with epitranscriptomic and transcriptomic tools, can unravel conserved regulatory mechanisms. CD Genomics supports similar studies by offering integrated genome assembly, methylation analysis, and functional genomics pipelines for plant epigenetics and beyond.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics
Journal: Viruses
Year: 2024
Genome sequence, antibiotic resistance genes, and plasmids in a monophasic variant of Salmonella typhimurium isolated from retail pork
Journal: Microbiology Resource Announcements
Year: 2024
Genes of Salmonella enterica Serovar Enteritidis Involved in Biofilm Formation
Journal: Applied Microbiology
Year: 2024
Complete genome sequence of the probiotic Bifidobacterium adolescentis strain iVS-1
Journal: Microbiology Resource Announcements
Year: 2023
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
