10x Genomics Single-Cell Sequencing
CD genomics is now offering single cell analysis utilizing various solutions of the 10X Genomics Chromium system, allowing the profiling of rare or heterogeneous populations of cells.
The Introduction of 10x Sequencing
Powerful approaches have become more accessible that allow the profiling of rare or heterogeneous populations of cells. The averaging that occurs in Conventional 'bulk' methods of sequencing does not allow the direct assessment of the fundamental biological unit—the cell—or the individual nuclei that package the genome. CD genomics provides access to the genomic and genetic analysis at single cell level using 10x genomics system and its engineered reagent delivery method. The droplet-based platforms Chromium 10x System, powered by GemCode Technology, enables the biomedical researchers and clinicians to make important new discoveries using this powerful approach as the technologies and tools needed for conducting single cell studies. Moreover, the droplet-based platforms, maximizing the throughput of cells and meanwhile downscaling the reactions to nanoliter volumes, has been shown to improve detection sensitivity and quantitative accuracy. The droplet-based methods hold advantages, allowing direct analysis of rare cell types or primary cells for which there may be insufficient material for conventional bulk sequencing protocols. It is also ideal to profile interesting subpopulations of cells from a larger heterogeneous population, for example, malignant tumor cells within a tumor mass, or hyper-responsive immune cells within a seemingly homogeneous group. It can even reveal entirely new cell types. 10X genomics also permits to study cellular states in scenarios such as embryonal development, cancer, myoblast and lung epithelium differentiation, and lymphocyte fate diversification.
Encapsulate your sample into hundreds to tens of thousands of uniquely addressable partitions in minutes, each containing an identifying barcode for downstream analysis. Each Gel Bead, infused with millions of barcoded oligonucleotides, is mixed with a sample, which can be high molecular weight (HMW) DNA, individual cells, nuclei, or Cell Beads. Gel Beads and samples are then added to an oil-surfactant solution to create Gel Beads in EMulsion (GEMs), which act as individual reaction vesicles in which the Gel Beads are dissolved and the sample is barcoded. Barcoded products are pooled for downstream reactions to create short-read sequencer compatible libraries. After sequencing, the resulting barcoded short read sequences are fed into turnkey analysis pipelines that use the barcode information to map reads back to their original HMW DNA, single cell, or single nucleus of origin.
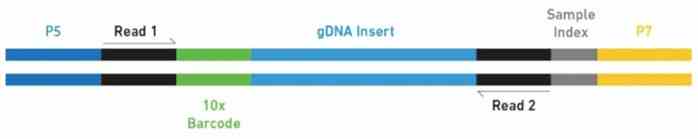 Figure 1. Schematic overview of the fragment of a final 10x Chromium Genome library
Figure 1. Schematic overview of the fragment of a final 10x Chromium Genome library
Chromium™ Single Cell Gene Expression Solution
Maximizing the throughput of cells is key to identify the cell types from complex tissues. The 10X Genomics Chromium Single Cell Gene Expression Solution can be used to perform transcriptome measurement in individual cells with sequencing large numbers of single cells can recapitulate bulk transcriptome complexity. CD Genomics is dedicated to providing high-quality single cell transcriptome profiling service using the 10X Genomics Chromium Single Cell Gene Expression Solution, along with turnkey software tools, which allows the creation of high complexity libraries from single cells to maximize insight from any sample type. The Cell Ranger analysis pipelines perform primary analysis and visualization. The Cell Ranger analysis pipelines perform standard analysis steps such as demultiplexing, alignment, and gene counting. Cell Ranger leverages the10x Barcodes to generate expression data with single-cell resolution. This data type enables applications including cell clustering, cell type classification, and differential gene expression at a scale of hundreds to tens of thousands of cells.
The Chromium™ System enables single cell transcriptional profiling of up to tens of thousands of cells (typically 1000-6000), by partitioning cells into nanoliter-scale GEMs, where all generated cDNA share a common 10x Barcode is used to associate individual reads back to the individual partitions. Incubation of the GEMs produces barcoded, full-length cDNA from poly-adenylated mRNA Single Cell 3' Library comprises standard Illumina paired-end constructs which begin and end with P5 and P7. 16 bp 10x Barcode and 12 bp molecular barcodes are encoded in Read 1, as the first base pairs of the library insert. Sample index sequences are incorporated as the i7 index read.
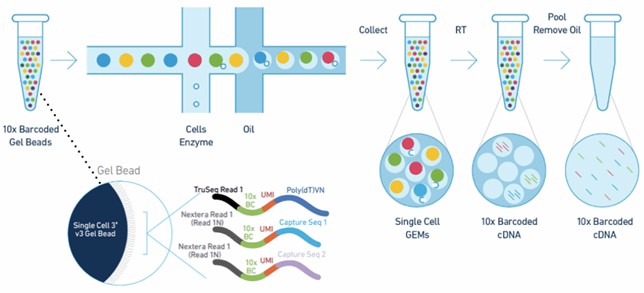 Figure 2. The Chromium™ cell partitioning workflow for scRNA sequencing
Figure 2. The Chromium™ cell partitioning workflow for scRNA sequencing
10x Genomics Single Cell Protocols require a suspension of viable single cell or single nucleus as input. Researchers must submit a dissociated suspension of live cells. To obtain high-quality data, when it is crucial to maximize viability and minimize the presence of nuclear aggregates, dead cells, cellular debris, cytoplasmic nucleic acids, and potential inhibitors of reverse transcription. Accurate cell counting is critical to application success. The single cell suspension between 700-1200 cells/ul is recommended for optimal target cell recovery. It is recommended that the cell suspension should be counted 3-4 times and the standard deviation between all counts should be < 25%.
- Feature barcoding: cell surface barcoding
Feature barcoding technology enables multiplxing and doublet detection and/or identifying cell-surface protein isoforms, detecting protein for low abundance transcripts, and further increasing phenotypic specificity.
- Feature barcoding: CRISPR screening
Implementing the CRISPR screening to conduct high-throughput and scalable functional genetic screens in hundreds to tens of thousands of single cells simultaneously.
Chromium™ Single Cell Immune Profiling Solution
The Chromium Single Cell Immune Profiling Solution is a comprehensive approach to understand the adaptive immune system of hundreds to tens of thousands of T and B cells in human or mouse with single cell resolution. CD genomics offers streamlined workflows that allow you to go from cells suspension to library prep, immune sequencing, and software analysis, enabling the assembly and annotation of full- length V(D)J segments and simultaneously assessment of TCR, Ig, and 5' gene expression in the same cell.
Single cell suspensions loaded onto the system are partitioned into GEMs, where transcripts are tagged with cells specific barcodes. The barcoded cDNA is then pooled for downstream processing and library preparation. For immune repertoire profiling, the cDNA undergoes targeted enrichment for T- or B-cell receptor transcripts prior to library preparation. The protocol produces Chromium Single Cell V(D)J libraries ready for Illumina sequencing.
For V(D)J enriched libraries, Read 1 encodes the 16 bp 10x™ Barcode, 10 bp molecular barcodes, and 13 bp Switch Oligo, as well as the 5' end of an enriched transcript. For 5' gene expression libraries, Read 1 encodes the 16 bp 10x Barcode and 10 bp molecular barcodes. Due to Enzymatic Fragmentation, for both libraries Read 2 encodes a random internal fragment of the corresponding insert. Sample index sequences are incorporated as the i7 index read. A schematic of the final library constructs is shown below.
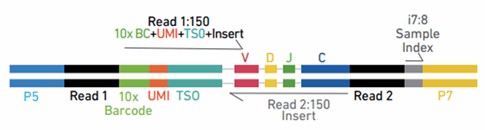 Figure 3. Chromium Single Cell V(D)J Enriched Library
Figure 3. Chromium Single Cell V(D)J Enriched Library
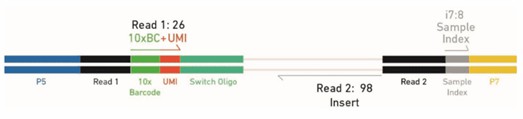 Figure 4. 5' Gene Expression Library Structure
Figure 4. 5' Gene Expression Library Structure
The Chromium™ linked-reads Sequencing Solution
The Chromium Genome Sequencing Solution enables molecular barcodes to tag reads that come from the same long DNA fragment, which provides the long-range information missing from standard approaches. As a result, we are able to link the short reads together by using a unique barcode to every short read generated from an individual molecule. Linked-reads can help access NGS dead zones, find more structural variants, resolve haplotypes and easily de novo genome assemblies.
For whole genome analyses, starting the process with high molecular weight (HMW) genomic DNA, sequencing-ready libraries of copies of unique barcoded gDNA is created. For whole exome analyses, by adding the Agilent SureSelectXT Human All Exon V6 Reagent Kit, the Target Enrichment libraries provide optimal exome application performance as well. Optimal performance has been characterized on input gDNA with a mean length greater than 65 kb, and this protocol outlines the extraction of HMW gDNA with optimal size from live cells.
The Chromium™ Single Cell Copy Number Profiling
CD genomics has the ability to sequence libraries from the DNA of single cells and analyze the genomic data, accurately detecting single cell CNV events.
Building on the 10x GEMs technology, The Chromium Single Cell CNV Solution is able to profile hundreds to thousands of cells. DNA from single cells is barcoded within the Cell Bead Gel Bead partitions and the barcoded fragments are then pooled for library production. The barcoded library fragments can be easily traced back to the cells from which they originated using downstream bioinformatics tools. It provides a comprehensive, scalable approach to determine genome heterogeneity and map clonal evolution by profiling hundreds to thousands of cells in a single sample. It is easier than ever to study complex pathogenesis, including cancer progression and genetic disorders, at unprecedented scale and resolution. Single cell CNV events will be called at around 2Mb resolution, but on clusters of cells, CNV events can be detected down to hundreds of Kb resolution.
The Chromium™ ATAC Sequencing
The 10x Chromium™ System designed single cell ATAC (Assay for Transposase Accessible Chromatin) solution to help understand the regulatory landscape of the genome. also enables understanding of epigenetic and regulatory variation across tens of thousands of cells with the Chromium Single Cell Assay for Transposase Accessible Chromatin (ATAC) Solution. Starting with hundreds to tens of thousands of nuclei, transposase enzyme is used to preferentially tag accessible DNA regions with sequencing adaptors. Transposed DNA fragments from individual nuclei are distinguished with one of ~750,000 possible 10x Barcodes. It profiles 500 to 10,000 nuclei per chanel and interrogate open chromatin profiles of individual nuclei.
Advantages of 10x Single-Cell Sequencing
- High Cellular Throughput: The microfluidic "double-cross" system features an 8-channel configuration, allowing up to 10,000 cells to be captured per channel. This enables the analysis of 50,000 to 80,000 sample cells in a single run across all eight channels.
- High Single-Cell Sequencing Coverage: The sequencing depth reaches an average of 50,000 reads per cell.
- High Cell Capture Efficiency: With a single-cell capture efficiency as high as 65%, the system can accurately identify rare cell types, facilitating research on rare samples or those with a small number of cells.
- True Single-Cell Sequencing: The probability of a single GEM (Gel Bead in EMulsion) capturing multiple cells is exceedingly low (less than 0.09% per 1,000 cells).
- Wide Cell Adaptability: The system imposes no restrictions on cell size (cells with diameters over 40 μm require preparation of cell nuclei) or cell type, accommodating tissue cells, immune cells, blood cells, cancer tissue cells, neural cells, and more.
Applications of 10x Single-Cell Sequencing
Immunological Studies
- Identification of Immune Cell Subtypes: Use single-cell technology to identify and label different subtypes of immune cells.
- Genetic Diversity Analysis: Analyze the genetic diversity of immune cells and the high heterogeneity caused by pathogens.
- Discovery of New Marker Genes: Identify rare immune cell types, discover new marker genes, and study the molecular mechanisms of immune responses.
Tumor Research
- Identification of Tumor Cell Subtypes: Recognize different cell subtypes within tumors, providing precise genetic and transcriptomic data.
- Tumor Heterogeneity Research: Study tumor cell heterogeneity, clustering, and the discovery of new cell types, seeking new pathogenic pathways and mechanisms.
Neural Development Research
- Neuron Heterogeneity Analysis: Investigate the heterogeneity and clustering of different neuron cells.
- Molecular Regulation Mechanism Analysis: Analyze the molecular regulation and differentiation mechanisms of neuron cells, helping to understand the mechanisms of neural diseases.
Brain Development Research
- Human Brain Gene Expression Atlas: Obtain a gene expression atlas of human brain cells, analyze cell heterogeneity and complex cell groups.
- Understanding Function and Mechanisms: Understand the normal function and abnormal mechanisms of the human brain.
Disease Typing
- Discovery of Abnormal Cell Types: Find abnormally proliferating cell types to assist in disease typing.
Stem Cell Differentiation
- Cell Heterogeneity Analysis: Analyze the heterogeneity of stem cells and identify cells of different phenotypes.
- Discovery of Specific Markers: Find specific markers of different types of stem cells and analyze the molecular mechanisms of stem cell development and differentiation.
Embryonic Cell Development Research
- Construction of Cell Lineages: Study cell lineage construction based on the characteristics of different cell subtypes to assist in disease typing.
Cell Atlas Construction
- Recognition of New Cell Types: Accurately identify various cell types through single-cell transcriptome analysis and discover new cell types and marker genes.
10x Single-Cell Sequencing Workflow
The comprehensive workflow for preparing single-cell libraries using 10x Genomics encompasses three major steps: the preparation of a single-cell suspension, the construction of GEMs (Gel Bead in EMulsion), and library construction. This is followed by cell lysis and reverse transcription, sequencing, and data analysis. CD Genomics ensures the 10x Genomics Single-Cell Sequencing process is executed with full integrity, precision, and efficiency.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
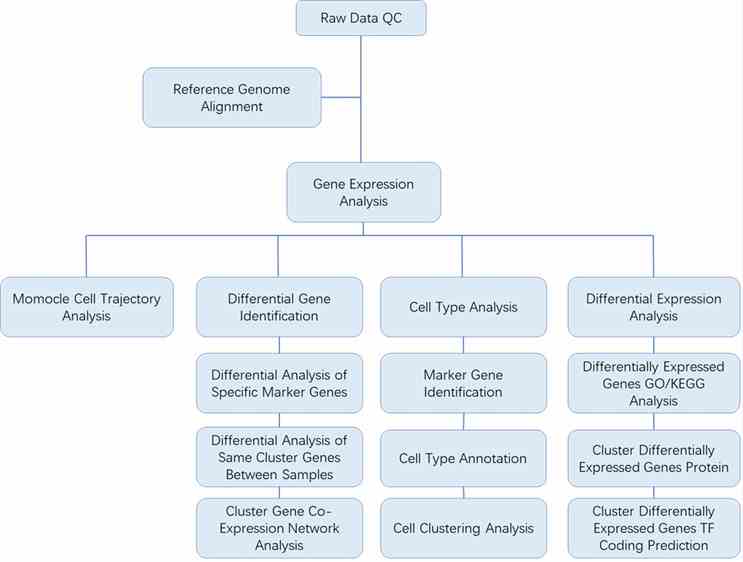
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in 10x Single-Cell Sequencing for your writing (customization)
If you have additional requirements or questions, please feel free to contact us.
Partial results are shown below:
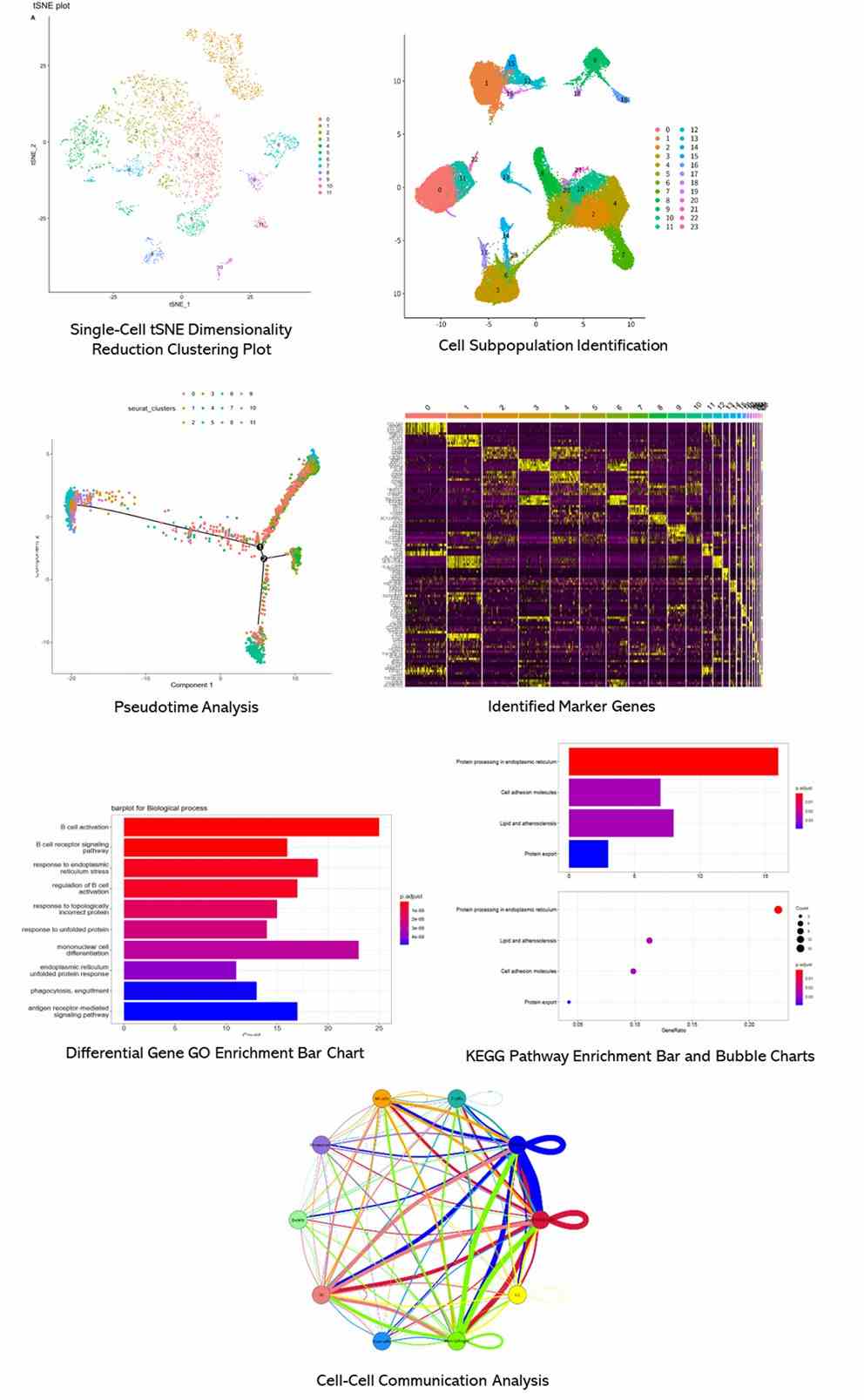
1. Why Conduct Single-Cell Sequencing Research?
Heterogeneity exists even among adjacent cells within the same tissue, such as brain or cardiac tissues. This is particularly pronounced in tumors, which are amalgamations of various mutated cells. Conventional bulk phase sequencing, which analyzes the genetic material from a mixture of cells within a tissue and its microenvironment, may obscure critical information due to cellular heterogeneity. In contrast, single-cell sequencing resolves this issue by significantly enhancing the resolution of sequencing analysis to the single-cell level. This precision allows for a more accurate understanding of genetic information, and it provides powerful insights into individual development and disease research.
From 2013 to 2020, single-cell sequencing technology has been consistently highlighted as a breakthrough annual technology by prestigious journals such as Science and Nature. This technique is steering a new wave of revolution in the field of biomedical research.
2. What Single-Cell Products Does 10x Genomics Currently Offer?
10x Genomics offers a suite of single-cell solutions based on their Chromium system, which utilizes the advanced 10x Next GEM technology. These solutions include:
- Single-Cell Transcriptomics: For gene expression analysis at the RNA level.
- Single-Cell CNV-Seq: For examining copy number variations (CNVs) at the DNA level.
- Feature Barcode Protein Detection: For profiling surface proteins using feature barcoding.
- Single-Cell Immune Profiling (V(D)J-Seq): For comprehensive immunological analysis, including the profiling of immune cells' antigen receptor repertoires.
- Single-Cell ATAC-Seq: For assessing chromatin accessibility and providing insights into the epigenetic landscape.
3. Is Single-Cell Sequencing Currently Feasible for Plants?
Unlike single-cell sequencing in animals, plant single-cell research is still in its infancy. One significant challenge stems from the presence of the plant cell wall, which necessitates the preparation of protoplasts before creating single-cell suspensions. This process is often complicated by issues related to enzymatic digestion times and buffer osmolarity. Additionally, the lower cellular heterogeneity within plant tissues complicates cell clustering efforts. Furthermore, the size of certain plant cells can be too large for compatibility with the 10x Genomics platform, making single-cell sequencing more challenging.
Single-Cell RNA Sequencing Resolves Molecular Relationships Among Individual Plant Cells
Journal: Plant physiology
Impact factor: 7.479
Published: 04 February 2019
Background
Different cell types in organisms result from varying gene expression, critical for multicellular functions. Single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of gene expression within cells, particularly in animals, revealing diverse patterns and rare types. Its application in plants faces challenges like isolating cells within the cell wall, yet recent studies in Arabidopsis roots using scRNA-seq provide insights into gene expression and development, showing promise for plant biology.
Materials & Methods
Sample Preparation
- Arabidopsi seeds
- Protoplast isolation
Sequencing
- scRNA-seq
- 0X Genomics Cell Ranger
- tSNE dimensionality reduction and clustering analysis
- Pseudotime trajectory analysis
- GO enrichment analysis
- Global analysis of cell differentiation status
- Heat map analysis of gene expression
Results
This study utilized the 10X Genomics Chromium platform to analyze 7522 single-cell transcriptomes from Arabidopsis root tips, revealing high reproducibility and identifying nine major cell clusters. Differential gene expression and GO analysis highlighted tissue-specific functions such as root hair and stele-related genes. Marker gene analysis enabled precise assignment of clusters to distinct root tissues. Subcluster analysis within the stele identified specific cell types like xylem and phloem, showcasing the dataset's ability to uncover rare cell subtypes. Gene expression gradients across clusters indicated developmental dynamics, illustrating a spatial differentiation pattern from meristematic to mature cells.
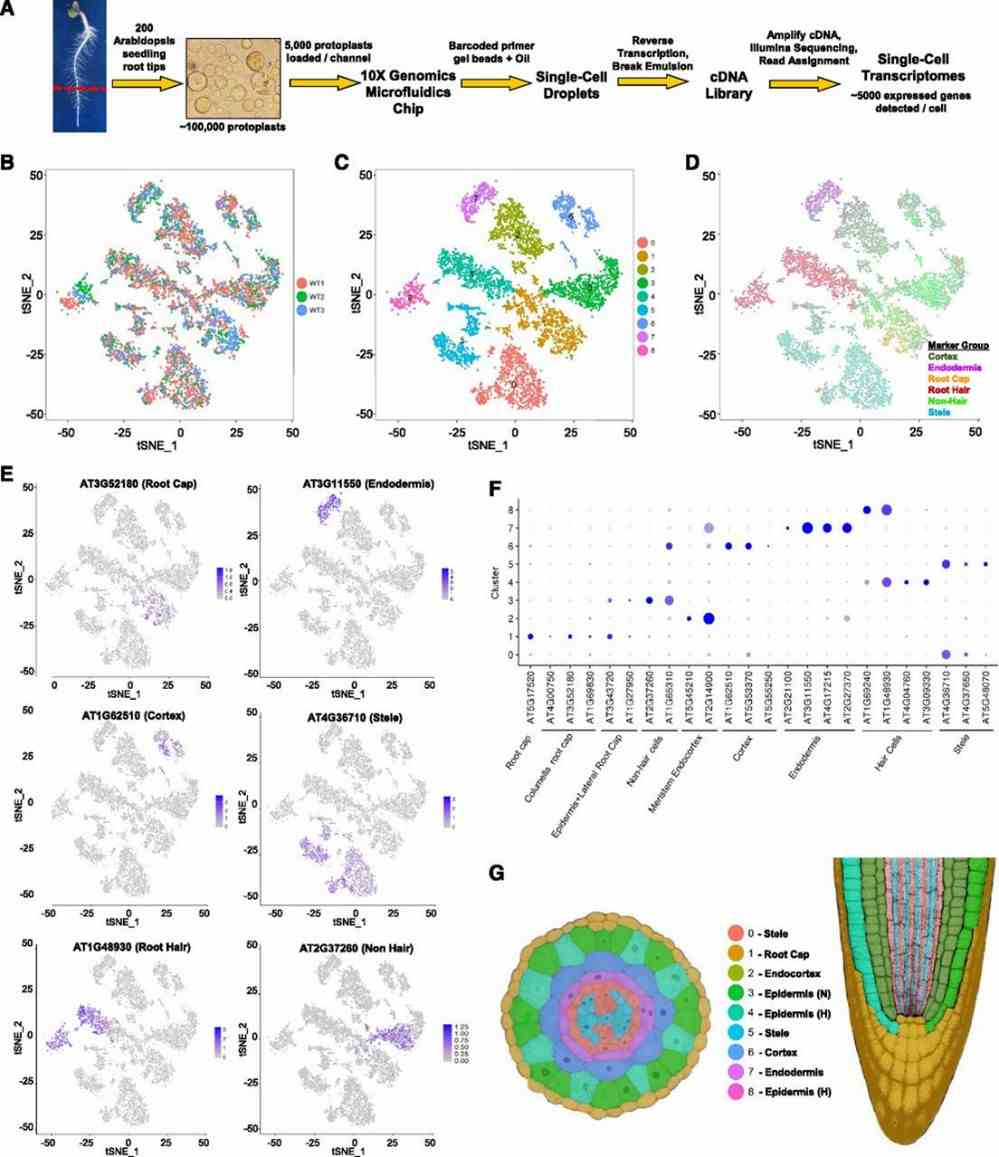 Figure 1. Isolation and cluster analysis of single-cell transcriptomes from wild-type Arabidopsis roots.
Figure 1. Isolation and cluster analysis of single-cell transcriptomes from wild-type Arabidopsis roots.
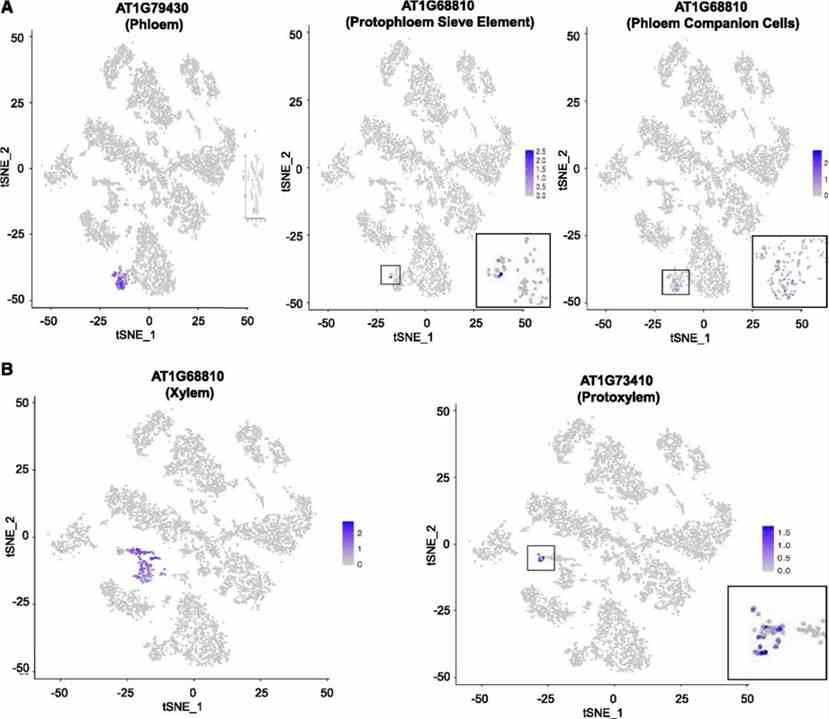 Figure 2. Stele marker gene expression on tSNE projection plots define clusters of distinct tissue and cell types in the stele.
Figure 2. Stele marker gene expression on tSNE projection plots define clusters of distinct tissue and cell types in the stele.
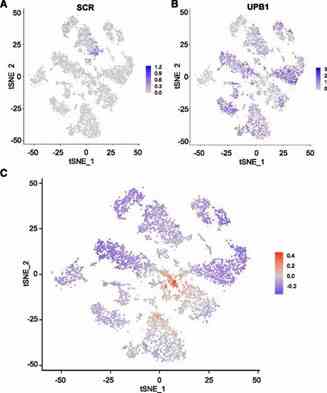 Figure 3. Intracluster developmental variation in gene expression.
Figure 3. Intracluster developmental variation in gene expression.
In this study, single-cell RNA sequencing (scRNA-seq) was employed to analyze the rhd6 and gl2 mutant root epidermis at high resolution. The analysis revealed distinct molecular phenotypes: rhd6 mutants lacked root-hair cells, with some cells adopting nonhair characteristics, while gl2 mutants lacked nonhair cells, with some cells expressing early nonhair markers. These findings underscore scRNA-seq's efficacy in identifying cell type-specific gene expression changes and characterizing mutant phenotypes at the single-cell level.
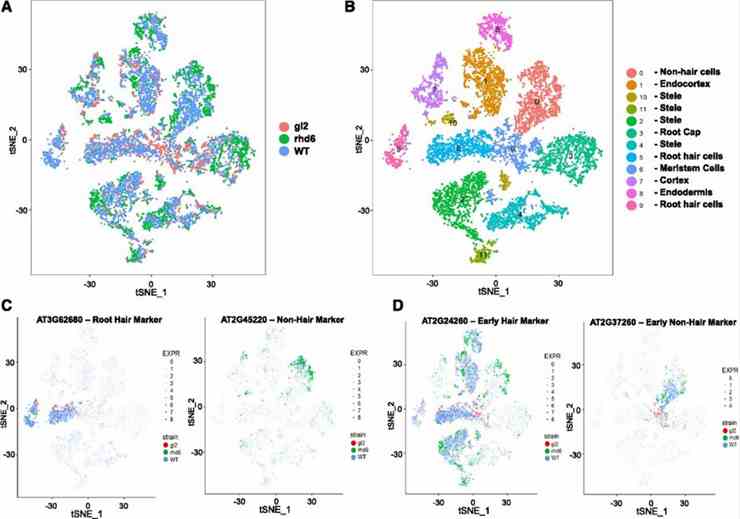 Figure 4. Comparative single-cell transcriptome analysis of wild-type and root epidermis mutant roots.
Figure 4. Comparative single-cell transcriptome analysis of wild-type and root epidermis mutant roots.
Conclusion
Single-cell RNA sequencing (scRNA-seq) has been less explored in plants compared to animals. The authors utilized a droplet-based microfluidics platform to perform high-throughput scRNA-seq on over 10,000 Arabidopsis root cells, revealing diverse tissue types and developmental stages. They identified rare cell types like quiescent center cells and traced developmental trajectories in root epidermal cells, including mutants, showcasing scRNA-seq's potential for detailed gene expression analysis in plants.
Reference
- Ryu KH, Huang L, Kang HM, et al. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant physiology. 2019, 179(4):1444-56.


 Sample Submission Guidelines
Sample Submission Guidelines
