Whole Genome SNP Genotyping
SNP, an abbreviation for Single Nucleotide Polymorphism, denotes a form of genetic polymorphism stemming from a mutation in the DNA sequence of the genome, precipitated by a solitary nucleotide alteration, which may involve the substitution, transversion, insertion, or deletion of a single base. In principle, SNPs can manifest as bi-allelic, tri-allelic, or tetra-allelic, yet, in practical applications, the latter two are extraordinarily rare and deemed inconsequential. Generally, when referencing SNPs, the focus tends to be on bi-allelic polymorphisms. These variations can manifest as transitions, exemplified by the conversion of a cytosine (C) to thymine (T), or its complementary strand counterpart, namely, the conversion of a guanine (G) to adenine (A). Alternatively, they may also manifest as transversions, such as cytosine to adenine (CA), guanine to thymine (GT), cytosine to guanine (CG), or adenine to thymine (AT). It is pertinent to note that transitions consistently exhibit markedly higher occurrence rates compared to other categories, with transition-type SNPs accounting for approximately two-thirds of the total.
Why Detect SNP Genotyping?
Common SNPs have the potential to manifest in both coding and non-coding regions within the genome. While their occurrence within coding regions is relatively infrequent, their capacity to influence gene functionality, consequently instigating alterations in biological traits, renders them of paramount significance in the realm of genetic disease investigation. As the third-generation genetic markers, SNPs are intricately distributed across the entire genomic landscapes of animals and humans alike. They exhibit a pronounced correlation with functional genes, boast minimal mutation rates, and exemplify robust genetic stability. Moreover, they lend themselves to high-throughput analysis and streamlined automation. It is noteworthy that certain SNP loci may not directly correlate with the expression of disease-associated genes. However, due to their proximity to particular pathogenic genes, they assume pivotal roles as crucial genetic markers in these contexts.
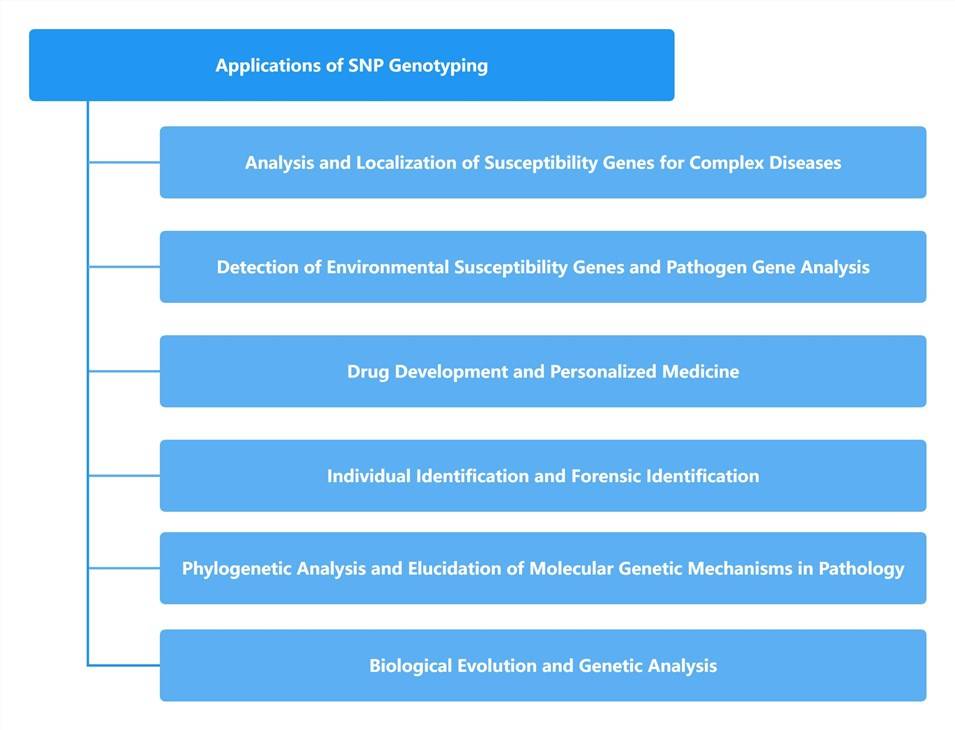
Microarray-based SNP Analysis
Gene chips offer a host of advantages, including substantial throughput, impeccable accuracy, and user-friendly operation, rendering them exceptionally fitting for applications where the specific target loci are pre-established. Particularly when the quantity of target loci surpasses a certain threshold, gene chips demonstrate a notable reduction in overall expenses coupled with heightened efficiency.
Gene chip technology is based on the principle of base complementary hybridization. It involves designing probe sequences for known SNP loci and labeling DNA probes with isotopes or fluorescent markers. These labeled DNA probes are hybridized with the target DNA to obtain biological information. Gene chips utilize microarray technology to integrate millions of probes onto a silicon chip or nylon membrane the size of a microscope slide. This allows the simultaneous analysis of 5,000 to 1,000,000 genotypes for an individual.
Our microarray-based SNP genotyping services comprise of Affymetrix SNP arrays and Illumina SNP arrays. The Affymetrix genotyping arrays on the GeneTitan instrument (Axiom array plates) and the GeneChip Scanner 3000 7G System (cartridge arrays) can be used for a variety of cases – for a few and a lot of samples from targeted to genome-wide applications by selecting the specific pre-designed genotyping arrays. The high density genotyping on Illumina Infinium or GoldenGate® arrays allow powerful genome-wide association studies (GWAS) and can accurately detect point mutations and copy number variants. Currently, Affymetrix offers genotyping arrays for livestock and aquaculture species (buffalo, cattle, chicken, pig, salmon and trout), crops (cotton, maize, soybean, strawberry and wheat), and biomedical model organisms (human, dog, mouse and Arabidopsis thaliana), while Illumina markets genotyping BeadArrays for human and non-human species (cattle, dog, maize, pig and sheep). SNP array has been taken as the preferred technique in some circumstances because of its high-density, assay accuracy, simple data analysis and easy data exchange between research programs. However, these commercially available SNP arrays or chips cannot be easily modified to suit individual experimental designs. In addition, relevant research cannot be conducted for species that do not have commercially available SNP arrays/chips.
The technical underpinnings of gene chips confer the subsequent advantages:
Elevated Throughput: Gene chips empower the simultaneous examination of an extensive array of genetic loci, with capacities extending to as many as 1,000,000 loci.
Stringent Standardization and Reproducibility: Each experimental run affords precise genetic information at predefined positions, eliminating inherent randomness and ensuring consistent outcomes.
Exemplary Accuracy: The chip production process is punctuated by rigorous quality assessments, with each detection locus subject to recurrent scrutiny, thus underpinning results of remarkable precision.
Efficiency: Experiments utilizing gene chips are expedited, with outcomes attainable in as little as 2 days, contributing to research expeditiousness.
Cost-Efficiency: Gene chips present a cost-effective solution, particularly conducive to large-scale, high-throughput genetic profiling endeavors.
Customizability: Gene chips can be tailored in accordance with specific target loci, with microarray chips offering enhanced flexibility for customization when juxtaposed with in situ synthesized counterparts.
NGS-based SNP Genotyping
NGS (Next-Generation Sequencing) technologies are powerful tools for SNP genotyping, because they can efficiently and accurately discover and genotype thousands of SNPs to investigate quantitative, functional and evolutionary genomics in human, animals and plants.
NGS is a DNA sequencing technology that has evolved from PCR and gene chips. In comparison to Sanger sequencing, NGS sequencing introduces reversible termination of the ends, allowing synthesis and sequencing to occur concurrently.
In NGS, individual DNA molecules must be amplified into clusters consisting of identical DNA compositions. These clusters are then synchronously replicated to enhance the fluorescence signal intensity, facilitating the reading of the DNA sequence. As read lengths increase, the cohesiveness of cluster replication decreases, leading to a decline in base sequencing quality. Therefore, NGS typically has shorter read lengths, usually not exceeding 500 base pairs. After sequencing, the information from fragmented DNA segments must be assembled, and the sequence assembly process has an error rate within the range of 0.1% to 15%, reducing accuracy.
Diminishing read lengths in NGS empowers the concurrent assessment of hundreds of thousands to even millions of gene sequences within a single sequencing run. This breakthrough effectively surmounts the constraints associated with low throughput that were characteristic of first-generation sequencing methods. A single NGS run can proficiently undertake multi-locus genotyping experiments, particularly when dealing with extensive sample sets, delivering notable time savings and cost reductions. Consequently, this technology has garnered extensive adoption in the realm of high-throughput gene sequencing.
There are various methods for Single Nucleotide Polymorphism (SNP) genotyping based on Next-Generation Sequencing.
Whole exome sequencing (WES): WES is a method that uses NGS to sequence only the exonic regions of the human genome, which encode proteins. While it is primarily used for studying the relationship between gene mutations and diseases, it can also be used for SNP genotyping because exons contain many SNP loci.
Whole genome sequencing (WGS): WGS involves sequencing all regions of the entire genome, including coding and non-coding regions. This method can be used to detect and genotype all SNPs in the genome, providing comprehensive genetic information.
Targeted Sequencing: The targeted sequencing approach entails the specific sequencing of genes or SNP loci of particular interest, allowing for the acquisition of highly focused SNP data. This methodology can be tailored to encompass the selective targeting of SNPs associated with specific diseases, enabling a meticulous examination of genetic variations pertinent to the pathology under investigation.
RNA Sequencing (RNA-Seq): RNA-Seq, while primarily harnessed for the investigation of gene expression profiles, offers a versatile utility encompassing the detection of Single Nucleotide Polymorphisms (SNPs). This multifaceted approach, predicated on the scrutiny of gene transcripts, facilitates the discernment of SNP expression and variations at the RNA level, thus affording a comprehensive understanding of genetic diversity and dynamics within this context.
Multiplexing: NGS technologies frequently enable the concurrent sequencing of multiple samples, leading to enhanced efficiency and cost reduction. This methodology proves highly advantageous in the context of large-scale single nucleotide polymorphism (SNP) genotyping endeavors, including Genome-Wide Association Studies (GWAS).
Genotyping by Sequencing (GBS): GBS is a high-throughput approach grounded in second-generation sequencing technology. This methodology facilitates the concurrent sequencing of DNA samples from multiple individuals and the determination of their respective genotypes. The GBS protocol encompasses library preparation, targeted sequencing of specific genomic regions achieved by DNA digestion utilizing restriction enzymes, and the use of high-throughput sequencers to produce extensive datasets rich in single nucleotide polymorphism (SNP) information.
dd-RAD (Double Digest Restriction-site Associated DNA sequencing): dd-RAD represents an additional SNP genotyping technique grounded in second-generation sequencing technology. This method entails the utilization of two distinct restriction enzymes to cleave DNA, subsequently followed by the sequencing of the cleaved products for the detection and genotyping of SNP loci.
2b-RAD: 2b-RAD is a reduced genome representation sequencing or restriction site associated DNA sequencing strategy, to discover and genotype genetic variants in a cost effective manner, which are applicable to both model organisms with known genomes and non-model organisms with unknown genomes. This technique is defined as the library construction by digesting DNA with restriction enzymes and analyzing the subsequent library with Illumina sequencing platforms.
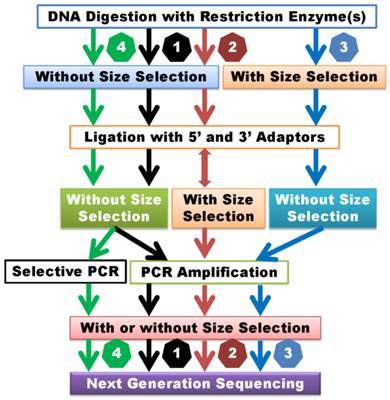
PCR-Based SNP Genotyping Techniques
PCR offers high sensitivity, low cost for scenarios with a limited number of loci to be tested, and shorter detection times, typically completed within 2-4 hours. It is operationally straightforward. However, it has limitations, such as the detection of a single locus, low throughput (usually between a few to a few hundred loci), and the inability to detect other mutations that may exist in DNA. False negatives and false positives are possible, making it suitable for scenarios with a small number of loci to test and lower throughput requirements.
Ligase Detection Reaction (LDR) Genotyping: This method primarily employs ligase detection reaction (LDR) technology. Taq DNA ligase is used to facilitate the reaction, which occurs only when two short nucleotide probes, one from each strand, are fully complementary to the target DNA sequence without any gaps. Detection is achieved through fluorescent scanning of fragment lengths, enabling SNP locus identification.
Multiplex SNaPshot SNP Genotyping: SNaPshot SNP genotyping is a method that simultaneously uses multiple PCRs to perform genetic typing of several known SNP loci. It utilizes DNA sequencing enzymes, four fluorescently labeled ddNTPs, extension primers of varying lengths adjacent to the polymorphic site, and PCR product templates. Primer extension terminates after a single base, and after gel electrophoresis on an ABI 3730 sequencer, the colors of peaks indicate the incorporated base, determining the sample's genotype and SNP locus corresponding to the extension product based on peak mobility.
Taqman Probe Method: This method involves adding two probes with different fluorescent labels to the PCR reaction system. Each probe fully complements one of the two alleles. Normally, due to the close proximity of the 5' end fluorescent group and 3' end quencher group on the probes, fluorescence is quenched. As PCR proceeds, the probe fully complementary to the template is gradually cleaved by Taq DNA polymerase 5'→3' exonuclease activity, leading to the separation of the 5' end fluorescent group from the 3' end quencher group, dequenching the fluorescence. Conversely, the probe representing the other allele, which does not perfectly match the template, is not efficiently cleaved, and no fluorescence is detected. SNP locus detection is achieved by measuring changes in fluorescence values using the corresponding instrumentation.
PCR-ARMS (Amplification Refractory Mutation System): ARMS is a PCR amplification method designed for specific SNPs. Through primer design, it selectively amplifies DNA fragments containing or not containing the specific SNP.
PCR-based techniques provide a range of options for SNP genotyping, each distinguished by its unique features and adaptability to specific research contexts.
Mass Spectrometry-Based SNP Genotyping
MassARRAY SNP Genotyping: Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) serves as the foundational technology for this method. Initially, PCR is employed to amplify gene segments containing SNPs. Subsequently, single-base extension is conducted using sequence-specific primers. Following this, the sample analyte is co-crystallized with the chip matrix and excited by a nanosecond (10-9s) laser pulse within a vacuum tube. This process results in the desorption of nucleic acid molecules, converting them into single-charged ions. Due to the inverse relationship between ion mass and ion flight time in an electric field, the precise molecular weight of the sample analyte is ascertained by measuring the flight time of nucleic acid molecules within the vacuum tube. As a result, SNP site information can be reliably detected. This method is well-suited for medium to high-throughput SNP analysis, particularly when dealing with more than ten SNP sites.
CD Genomics is committed to providing SNP genotyping on a genome-wide scale and an affordable way. Our experienced scientists can offer professional supports for your project to meet your research objectives and budget.
Comparison of SNP Genotyping & RAD-Seq Methods
Here's a clear side-by-side overview of the genotyping and RAD-seq techniques:
| Method | Genome-wide | Fragment Source | Size Selection | Throughput | Best Use Case |
|---|---|---|---|---|---|
| RAD-seq (classic) | ✅ Yes | Single enzyme + random shearing | Physical shearing | Medium | Non-model species without reference genome |
| ddRAD-seq | ✅ Yes | Two enzymes + size-selected fragments | Gel-based, precise range | High | Balanced SNP discovery with uniform coverage |
| 2b-RAD | ✅ Yes | Type IIB enzyme → uniform 33–36 bp tags | No size selection needed | Very High | High-density SNP mapping, even on degraded DNA |
| PCR-LDR Genotyping | ❌ Targeted only | PCR + ligase detection | N/A | Low | Validation of known SNPs for one or few loci |
| MassARRAY Genotyping | ❌ Targeted only | PCR + single-base extension + MALDI-TOF MS | PCR-based primers | Medium–High | Multiplex validation of ~10–100 SNPs |
Choose a method based on:
- 2b-RAD or ddRAD-seq for genome-wide discovery
- Classic RAD-seq for non-reference species
- PCR-LDR for small-scale, high-accuracy SNP tests
- MassARRAY for medium-throughput, multiplexed validation
Use of biostimulants for water stress mitigation in two durum wheat (Triticum durum Desf.) genotypes with different drought tolerance
Plant Stress | 2024The Restriction–Modification Systems of Clostridium carboxidivorans P7
Microorganisms | 2023In the land of the blind: Exceptional subterranean speciation of cryptic troglobitic spiders of the genus Tegenaria (Araneae: Agelenidae) in Israel
Molecular Phylogenetics and Evolution | 2023Genetic Modifiers of Oral Nicotine Consumption in Chrna5 Null Mutant Mice
Front. Psychiatry | 2021

 Sample Submission Guidelines
Sample Submission Guidelines
