AAV Genome Sequencing
What is the AAV Genome
The AAV (Adeno-Associated Virus) genome is a small but highly functional single-stranded DNA (ssDNA) molecule, spanning about 4.7 kilobases (kb). This compact genetic package is ingeniously designed with several key features:
- Inverted Terminal Repeats (ITRs): Located at both ends of the genome, these repetitive sequences are crucial for the virus's lifecycle. They form distinctive hairpin structures that are vital for the replication process and the packaging of the viral genome. These ITRs also play a key role in integrating the viral DNA into the host cell's genome.
- Rep Genes: The Rep (Replication) genes code for proteins that are essential for replicating the AAV genome and regulating its lifecycle. These proteins are involved in making copies of the viral DNA and can also assist in integrating the virus's genetic material into the host genome, ensuring the virus can persist and spread effectively.
- Cap Genes: The Cap genes produce the proteins that constitute the virus's protective outer shell, or capsid. These capsid proteins are not just structural components; they determine the virus's ability to infect specific types of cells. They also contribute to the virus's stability and its efficiency in delivering its genetic payload.
- Regulatory Regions: These regions manage the expression of the Rep and Cap genes. They act like traffic controllers, ensuring that the production of viral proteins occurs at the right time and in the right amounts throughout the virus's lifecycle.
What is NGS Sequencing of AAV
NGS (Next-Generation Sequencing) of AAV offers a deep dive into the viral genome, revealing a wealth of information about its structure and potential issues. This state-of-the-art technology meticulously maps the entire AAV genome, identifying genetic variants, mutations, and possible contaminants. Such detailed insights are crucial for fine-tuning AAV vectors used in gene therapy, ensuring they are both safe and effective. Moreover, NGS helps determine the exact concentration of AAV in samples and confirms the purity of vector preparations, ensuring they are free from unwanted viral or bacterial elements. In essence, NGS is a vital tool that supports the precise development, rigorous quality control, and successful application of AAV-based gene therapies.
Introduction to AAV Genome Sequencing
AAVs are currently one of the most widely utilized and significant vectors for in vivo gene therapy delivery. These vectors can deliver genetic payloads of up to 4.7 kb and can infect a diverse array of cell types with minimal pathogenicity. The production of AAVs begins with the construction of plasmids and concludes with the packaging into viral particles. Due to the structural characteristics of the AAV genome, producing AAV viruses with high-fidelity DNA is inherently challenging. To address these complexities, CD Genomics offers comprehensive AAV genome sequencing and quality control solutions designed to optimize the quality of AAV vectors.
Agarose gel electrophoresis, PCR/ddPCR, and Southern blotting have been extensively utilized for the assessment of AAV genome size and the identification of specific regions. However, these methods are limited in their capacity to provide comprehensive sequence information and insights into genomic integrity. Third-generation sequencing techniques, notable for their long-read capabilities, have been developed as potential solutions for AAV genome integrity analysis. Nevertheless, the intrinsic nature of the AAV genome as single-stranded DNA (ssDNA) necessitates its conversion to double-stranded DNA before library preparation for third-generation sequencing.
This conversion process, coupled with the modified termini present in AAV genomes, impedes precise terminal sequence analysis. Additionally, fragmented AAV genomes, being shorter than their intact counterparts, exhibit a significant bias during library preparation and sequencing, resulting in a disproportionate representation in sequencing datasets. Consequently, the analysis of AAV genome integrity using third-generation sequencing often yields results that underestimate the actual integrity, leading to data distortion. This distortion has severe implications for subsequent compliance reviews and the drug approval process. Therefore, while third-generation sequencing is suitable for accurate sequence analysis of AAV genomes, it is inadequate for the evaluation of terminal sequences and overall genome integrity.
In light of increasing regulatory demands, robust detection protocols and analytical methods are essential to ensure the safety and compliance of AAVs. CD Genomics has launched AAV genome sequencing and quality control services based on high-throughput sequencing technologies. By employing second-generation (NGS) and third-generation (Single-Molecule Real-Time Sequencing) sequencing analyses, we examine AAV purity, ITR sequence integrity, and target sequence accuracy. Our services are tailored to assist gene therapy drug development clients in enhancing their AAV production processes, thereby safeguarding the safety of subsequent clinical trials.
Advantages of Our AAV Genome Sequencing Service
- Accurate Results: Through precise sequencing methodologies, we achieve complete sequencing of the ITR regions, efficiently and accurately identifying mutation sites within these sequences.
- Rapid Sequencing: Our approach ensures that the sequencing of ITR regions is accomplished swiftly, resulting in expedited turnaround times for the delivery of results.
- Comprehensive Consultation Services: CD Genomics offers continuous technical support throughout the project lifecycle, whether during the initial, intermediate, or final stages.
- Compliance-Driven ITR Sequencing: CD Genomics operates fully accredited laboratories equipped with state-of-the-art instruments, adhering strictly to quality management control systems and standardized operating procedures (SOPs).
Applications of AAV Genome Sequencing
Gene Therapy Research
- Optimization of AAV Vectors
- Evaluation of Therapeutic Outcomes
Genome Editing
- Efficiency Analysis
- Specificity Validation
Vaccine Development
- Delivery System Optimization
- Immunogenicity Assessment
Basic Research
- Infection Mechanism Exploration
- Impact of Viral Variants
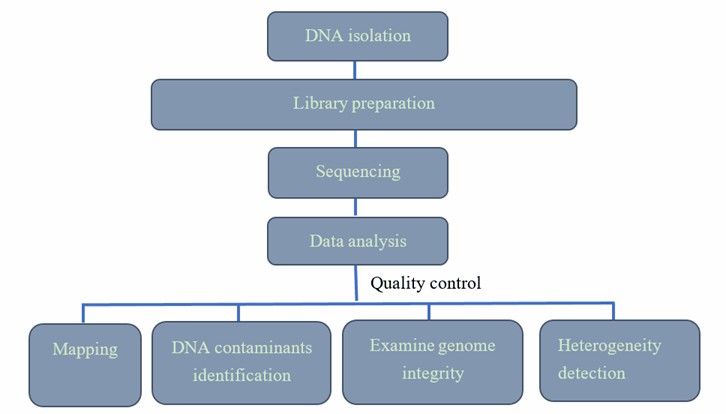
AAV Genome Sequencing Workflow
Our highly experienced expert team executes quality management following every procedure to ensure comprehensive and accurate results. Our AAV sequencing workflow is outlined below, including library prep, sequencing, and bioinformatics analysis.

Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis
We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in AAV Genome Sequencing for your writing (customization)
CD Genomics employs short-read whole-genome sequencing to conduct AAV genome sequencing, integrating Sanger sequencing and high-throughput platforms for comprehensive assessments. This approach enables the examination of genome integrity, the detection of heterogeneity, and the identification of DNA contaminants in AAV library preparations. We are dedicated to leveraging our extensive expertise and state-of-the-art technology to deliver superior services and high-quality products tailored to our clients' specific needs.
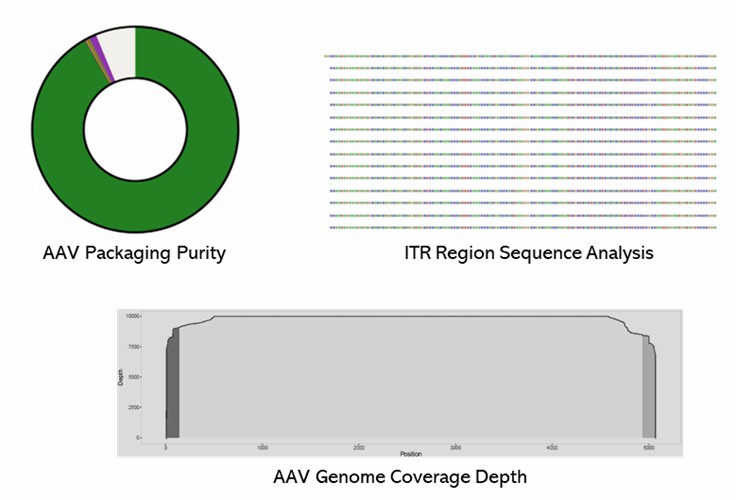
Demo Results
Partial results are shown below:
AAV Genome Seq FAQs
1. Why Choose AAV for Gene Therapy?
AAV stands out in gene therapy due to its high safety profile, low immunogenicity, physical stability, broad cellular tropism, and prolonged in vivo expression. These attributes make AAV one of the most commonly employed vectors in the field. The construction of AAV-based vectors is a cornerstone of gene therapy technology.
Researchers have undertaken extensive studies to optimize AAV vectors. These efforts include modifying the viral vector's genomic structure, increasing its cargo capacity, enhancing viral yield, and improving transduction efficiency across various tissue types. Such modifications aim to tailor AAV vectors to meet the specific needs of different diseases.
2. Why is the Cost of AAV so High?
- Purity Requirements for AAV Vectors
AAV vectors must meet stringent purity standards. Empty viral capsids can trigger immune responses and compete with fully encapsulated vectors for patient cell infection, necessitating higher doses to achieve therapeutic efficacy. This escalation in dosage not only raises the risk of high-dose side effects but also increases potential hepatotoxicity and carcinogenicity. Consequently, conducting thorough purity validation post-AAV vector construction becomes an essential step.
- High Mutation Rates in Specific ITR Structures
The AAV genome is flanked at both ends by inverted terminal repeat sequences (ITRs), which are crucial for rAAV packaging and replication. However, the ITR regions in AAV plasmids are unstable due to their palindromic sequences and high GC content, making them prone to mutations or deletions during bacterial replication. Even with the most standardized experimental protocols, AAV plasmids can exhibit mutation rates ranging from 5% to 15%.
- Challenges in ITR Detection
Given the high frequency of ITR mutations, assessing the integrity of the ITR regions in plasmids before viral packaging is critical. However, these regions are replete with palindromic sequences capable of forming stable secondary structures. During Sanger sequencing, these stable hairpin loops can inhibit PCR amplification, leading to sequencing interruptions and complicating sequence validation. Similarly, the conventional restriction enzyme method using SmaI to verify ITR integrity can be inadequate, as minor deletions may escape detection through gel electrophoresis.
3. What are the advantages of using AAV as a vector for gene therapy?
AAV has several advantages, including low immunogenicity, a broad host range, stable physicochemical properties, and long-term expression of exogenous genes.
Specifically, these advantages are:
- High Safety Profile: AAV is generally non-pathogenic to humans or may only cause very mild symptoms.
- Targeted Integration: AAV can integrate specifically into the AAVS1 site on human chromosome 19, which reduces the risk of insertional mutagenesis linked to random integration seen with other viral vectors.
- Sustained Gene Expression: AAV-mediated gene transfer systems facilitate persistent and stable expression of the introduced genes, which can be regulated by surrounding genetic elements.
- Broad Host Range: AAV can infect a wide variety of cell types, including both dividing and non-dividing cells.
- Thermal and Chemical Stability: The AAV transfer system demonstrates significant thermal stability and resistance to acidic and alkaline conditions, as well as organic solvents, making it convenient for storage.
AAV Genome Seq Case Studies
Adeno-associated Virus Genome Population Sequencing Achieves Full Vector Genome Resolution and Reveals Human-Vector Chimeras
Journal: Molecular Therapy Methods & Clinical Development
Impact factor: 4.771
Published: February 27, 2018
Background
Recombinant adeno-associated viruses (rAAVs) require precise quality control for gene therapy. Traditional methods like qPCR and gel electrophoresis don't fully capture genome fragmentation or contamination. Single-molecule real-time (SMRT) sequencing provides a detailed view of both full-length and truncated rAAV genomes, revealing errors in genome packaging and contamination from host-cell or viral fragments. This advanced technique offers a comprehensive analysis of rAAV vector populations.
Materials & Methods
Sample Preparation
- Purified viral vectors
- DNA extraction
Sequencing
- Library preparation
- SMRT Sequencing
- Pacific Biosciences RSII
- Read processing
- Alignment
- Circos plots, venn diagrams visualization
- Read count normalization
Results
AAV-GPseq uses SMRT sequencing to analyze full-length scAAV genomes from 5' to 3' ITRs. It reveals that shRNA cassettes lead to more truncated genomes and allows detailed characterization of ITR orientations and plus/minus strand ratios, providing insights into AAV vector packaging and replication dynamics.
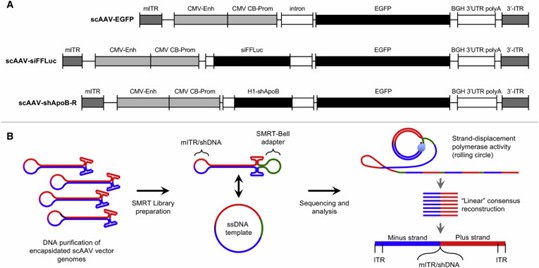 Figure 1. Single-Particle Resolution Profiling of scAAV Genomes by AAV-Gpseq.
Figure 1. Single-Particle Resolution Profiling of scAAV Genomes by AAV-Gpseq.
AAV-GPseq reveals that most vector genome reads are shorter than 500 bp, differing from gel analysis. By using spike-in DNA for normalization, it accurately quantifies full-length versus truncated genomes. This method shows that full-length vectors are less prevalent than traditional gel results suggest.
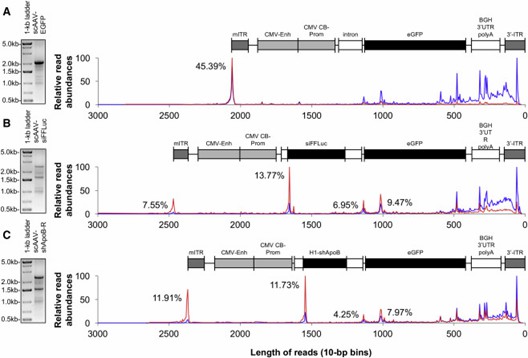 Figure 2. Abundance Assessment of Heterogeneous AAV Genome Populations.
Figure 2. Abundance Assessment of Heterogeneous AAV Genome Populations.
AAV-GPseq detects plasmid backbone sequences in vector genomes by revealing reads that extend beyond the ITR regions. This method identifies reads that align with plasmid backbone sequences, suggesting the presence of reverse-packaged or larger-than-unit-length genomes. The analysis also confirms that many short genomes, under 500 bp, include ITR sequences but are not present at detectable levels in purified vector samples.
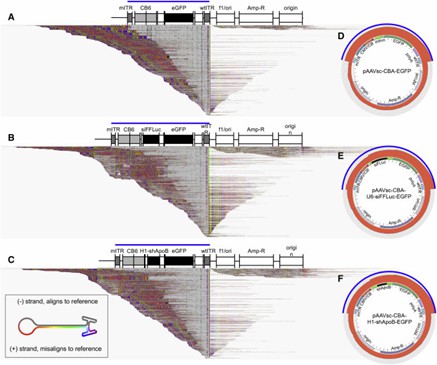 Figure 3. Alignments of Heterogeneous Vector Populations to the pCis-Plasmid Reference.
Figure 3. Alignments of Heterogeneous Vector Populations to the pCis-Plasmid Reference.
Conclusion
Traditional methods for assessing rAAV vector integrity are limited in profiling individual genomes. AAV-GPseq advances this by analyzing genomes from ITR-to-ITR, revealing chimeric sequences and plasmid backbones. Despite its strengths, challenges include underrepresentation of longer chimeric sequences and limitations with single-stranded AAVs. Overall, AAV-GPseq enhances the quality and safety evaluation of gene therapy vectors.
Reference
- Tai P W L, Xie J, Fong K, et al. Adeno-associated virus genome population sequencing achieves full vector genome resolution and reveals human-vector chimeras. Molecular Therapy Methods & Clinical Development, 2018, 9: 130-141.
Related Publications
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
