Introduction to Microsatellite and Microsatellite Genotyping
What are microsatellites?
Microsatellites, also known as short tandem repeats (STRs) or simple sequence repeats (SSRs), are tandem repetitive sequences in which the repeating unit contains from one to six nucleotides. The number of repeats is very variable in populations and within the alleles of an individual since microsatellites are prone to slippage during DNA replication. Despite the microsatellite instability occurring in all dividing cells, microsatellites remain stable in length due to the MMR system, which plays a major role in repairing DNA replication errors and regulating recombination events.
Microsatellites: informative genetic markers
Microsatellites are important genetic markers because they tend to be highly polymorphic, and they have been used as sex-specific markers, population-specific markers, etc. With the advent of PCR, microsatellite marker polymorphisms have replaced RFLPs as the markers for constructing linkage maps. The flanking regions are critical since they allow researchers to design locus-specific primers to amplify the microsatellites via PCR. That is, the primers for PCR are the sequences from the unique flanking regions. By developing a forward and a reverse primer on both sides of the microsatellite, we are able to amplify a locus-specific microsatellite region to determine the microsatellite variation.
Advantages of microsatellites as genetic markers:
- Highly polymorphic. The reason seems to be slippage replication.
- Locus-specific. Locus-specific primers allow us to amplify locus-specific microsatellites.
- Codominant (in contrast to RAPDs and AFLPs which are “binary, 0/1”).
- PCR-based. We work on tiny amounts of DNA with inexpensive laboratory instruments.
- A range of scales from individual ID to fine-scale phylogenies
Applications of microsatellite markers
Microsatellite markers are very useful (i) in the studies of biological diversity measured on the basis of genetic distance; (ii) to estimate gene flow and crossing over rates; (iii) to infer infraspecific genetic relationships; (iv) to construct linkage maps and define cultivar DNA fingerprints; (v) to map loci involved in quantitative traits; (vi) to determine the degree of kinship; (vii) in breeding using marker-assisted selection. That is, these markers are specifically useful in studies of population structure, genetic mapping, and evolution.
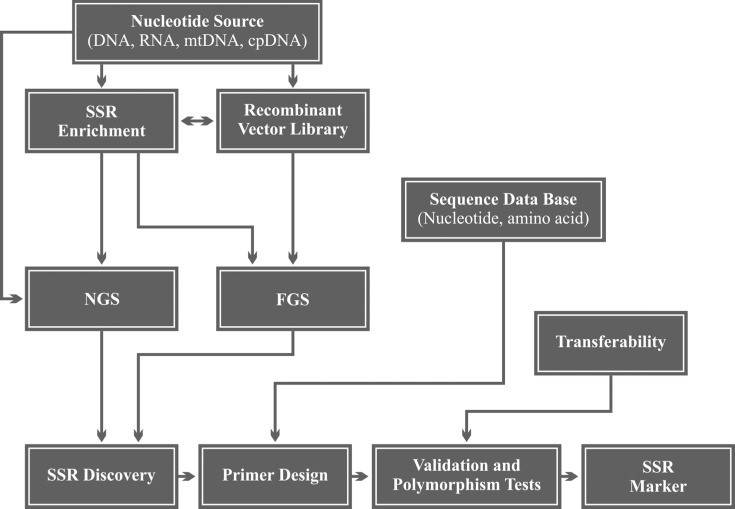 Figure 1. Workflow steps of SSR (microsatellite) marker development.
Figure 1. Workflow steps of SSR (microsatellite) marker development.
Microsatellite genotyping
Genotyping is the process to identify the genotype of each microsatellite. The workflow of microsatellite genotyping generally involves specific primer design, amplification of microsatellites, and Polymorphism testing.
- Specific primer design
If someone has developed SSR primers for a closely related species, these primers will be well worth checking in your species. If, however, there are no primers developed for related species, you may need to design your own. It is necessary to have the nucleotide sequences first, either from your own sequencing project or from the NCBI nucleotide database. SSR motifs can be identified using the MIcroSAtellite identification (MISA) tool (http://pgrc.ipk-gatersleben.de/misa). Locus-specific SSR primers can be designed with SSRLocator v1.0.
- Amplification of microsatellite
Once the SSR primers have been developed, it is necessary to validate these primers using PCR amplification. Each 20 uL SSR-PCR reaction mixture consists of DNA polymerase, 1×reaction buffer, 2 mmol L-1 MgCl2, 40 umol L-1 dNTP mix, 0.5 umol L-1 forward primer, 0.5 umol L-1 reverse primer, and 50 ng genomic DNA. Thermal cycling is performed in a PCR instrument under the following conditions: initial denaturation at 94°C for 5 min, 36 cycles of denaturation at 94°C for 50s, annealing at optimal temperature for 1 min, and extension at 72°C for 1 min, as well as a final extension at 72°C for 7 min.
- Polymorphism testing
The PCR results can be visualized by agarose gel electrophoresis or polyacrylamide gel electrophoresis (PAGE). PAGE genotyping is labor intensive but provides good resolution. For fine-scale studies, marked SSR primers can be synthesized with a fluorescence dye for genotyping by capillary electrophoresis using instruments such as the ABI 3730 DNA Analyzer. In this case, each PCR product is loaded into a capillary containing a polyacrylamide matrix in which the electrophoresis is performed. The emitted fluorescence is captured and the molecular mass of the PCR product is determined. An electropherogram is then generated which shows luminescence peaks corresponding to each amplified allele.
References:
- Vieira M L C, Santini L, Diniz A L, et al. Microsatellite markers: what they mean and why they are so useful. Genetics and molecular biology, 2016, 39(3): 312-328.
- Saha D, Rana R S, Chakraborty S, et al. Development of a set of SSR markers for genetic polymorphism detection and interspecific hybrid jute breeding. The Crop Journal, 2017, 5(5): 416-429.
- Guichoux E, Lagache L, Wagner S, et al. Current trends in microsatellite genotyping. Molecular ecology resources, 2011, 11(4): 591-611.


 Sample Submission Guidelines
Sample Submission Guidelines
