Sanger Sequencing vs. Next-Generation Sequencing (NGS)
Introduction
Within the sphere of DNA sequencing, two groundbreaking methodologies - Sanger Sequencing and Next-Generation Sequencing (NGS) - have transformed the biological sciences. Each of these techniques operates based upon distinct principles and are applied to a variety of uniquely fitting roles in research. Both come with their individual advantages and restrictions. Comprehending the disparities between Sanger sequencing and NGS is vital for scientists and clinicians, as it heavily informs the layout of experimental design, interpretation of derived data, and eventually the overall output of research conducted. This discursive piece embarks upon an explorative comparison of Sanger sequencing against NGS, examining their respective benefits, the limitations they pose, and the context in which these applications are suitably employed.
Differences Between NGS and Sanger Sequencing
Principle of Operation
Sanger Sequencing, recognized as capillary electrophoresis sequencing, operates by integrating fluorescently tagged dideoxynucleotides (ddNTPs) during DNA synthesis. Each ddNTP halts the elongation of the DNA strand at precise nucleotide locations, facilitating DNA sequence determination through capillary electrophoresis.
In contrast, NGS utilizes a diverse array of mechanisms, including reversible terminator chemistry, real-time single-molecule sequencing, and nanopore-based sequencing, to accomplish high-throughput sequencing.
Throughput and Scalability
The hallmark of NGS lies in its notable throughput and scalability. NGS platforms exhibit the capacity to sequence a multitude of DNA fragments within a single operational cycle, thereby facilitating extensive genomic coverage and concurrent analysis spanning hundreds to thousands of genes or gene regions. This attribute markedly outpaces the constrained throughput inherent in Sanger sequencing, which conventionally processes DNA fragments serially, one at a time.
Sensitivity and Detection Limit
NGS offers superior sensitivity compared to Sanger Sequencing, particularly in detecting low-frequency variants and rare mutations. The deep sequencing capacity of NGS allows for the identification of subtle genetic variations present at frequencies as low as 1%. In contrast, Sanger sequencing may exhibit lower sensitivity, with a detection limit typically around 15-20%.
Discovery Power
NGS distinguishes itself in the revelation of novel variants and genetic revelations attributable to its high-throughput capacity and expansive genome coverage. Scholars harnessing NGS technology can unveil hitherto unnoticed genetic variations and attain a more profound comprehension of genetic intricacies and diversity. Although Sanger Sequencing upholds precision and dependability, it may fall short in the realm of discovery potency afforded by NGS platforms.
You may interested in
Learn More
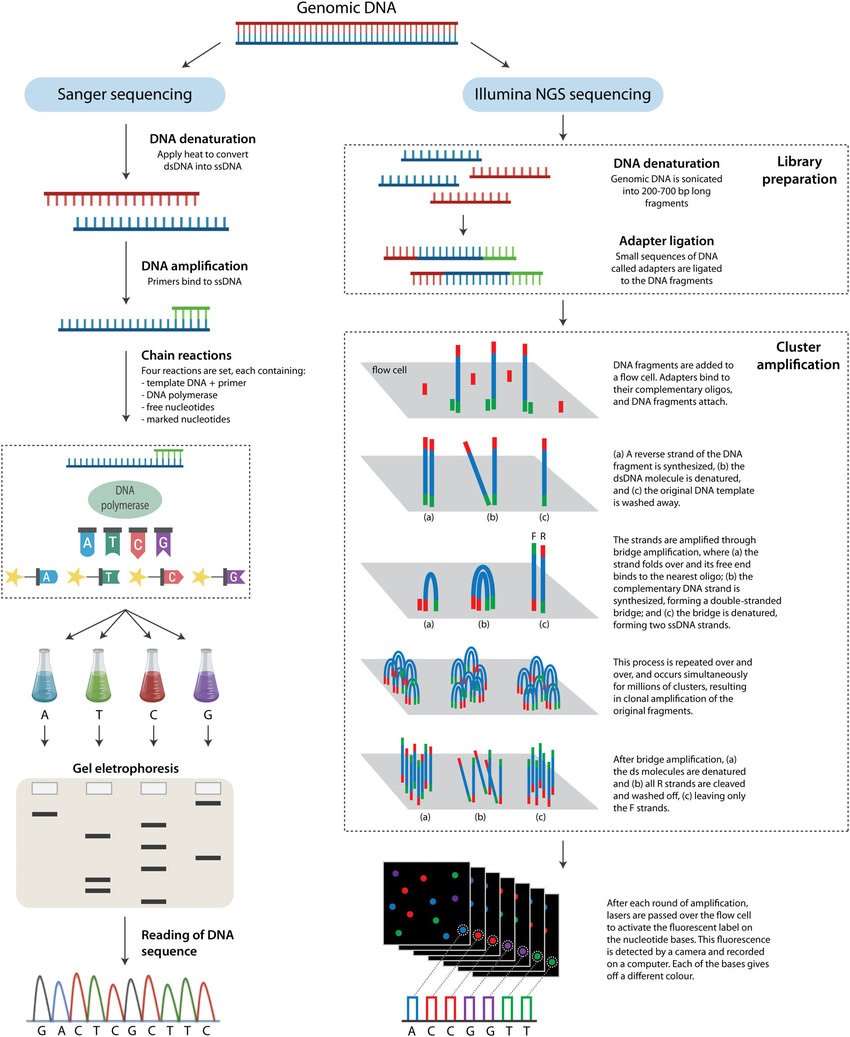 Comparison between Sanger sequencing and NGS technologies.
Comparison between Sanger sequencing and NGS technologies.
Benefits of Sanger vs Illumina
Fast and Cost-Effective Sequencing for Low-Number Targets:
Sanger Sequencing remains a viable option for sequencing a limited number of targets, such as single genes or short DNA fragments. Its familiarity, robustness, and cost-effectiveness make it suitable for small-scale sequencing projects requiring high accuracy and specificity. Additionally, Sanger sequencing can be performed without the need for complex library preparation steps, offering a straightforward workflow.
Familiar Workflow and Higher Sequencing Depth:
Sanger Sequencing presents a workflow familiar to many research laboratories, firmly established over time. Researchers adept in Sanger sequencing can harness its benefits for the targeted sequencing of precise genomic regions or genes. Furthermore, Sanger sequencing can attain enhanced sequencing depth, thereby facilitating heightened sensitivity in the detection of variants occurring at low frequencies.
Learn More
Limitations of Sanger vs Illumina
Limited Throughput and Scalability:
Despite its notable accuracy and reliability, Sanger Sequencing is hampered by diminished throughput and scalability in contrast to Next-Generation Sequencing (NGS) technologies. The labor-intensive protocols intrinsic to Sanger sequencing confine its utility in large-scale sequencing endeavors necessitating elevated sample throughput. Moreover, with the escalation in the number of targets, the cost-effectiveness of Sanger sequencing wanes, rendering it less conducive for comprehensive genomic analyses.
Elevated Costs for High-Volume Targets:
Sanger Sequencing may incur exorbitant expenses when tasked with sequencing a substantial quantity of targets (>20), as each target typically mandates individual sequencing reactions. The cumulative expenditure encompassing reagents, labor, and instrumentation associated with Sanger sequencing may outweigh its virtues of precision and established methodology, particularly within high-throughput environments.
Comparison of Sanger Sequencing and NGS
Sanger Sequencing and NGS embody divergent methodologies in DNA sequencing, each characterized by distinct advantages and limitations. Discerning the relative merits and drawbacks of these approaches is paramount for the judicious selection of sequencing strategies tailored to specific research or clinical requisites.
| Aspect | Sanger Sequencing | NGS |
| Benefits | - Demonstrates high accuracy and reliability | - Exhibits high throughput and scalability |
| - Embraces a familiar workflow with established protocols | - Offers superior sensitivity for low-frequency variants | |
| - Provides higher sequencing depth for variant detection | - Encompasses comprehensive genomic coverage | |
| Limitations | - Exhibits lower throughput, sequencing one fragment at a time | - Presents initial setup and data analysis complexities |
| - Incurs higher costs for high-volume targets | - Entails cost considerations for small-scale projects | |
| - Demonstrates limited discovery power compared to NGS | - Carries a potential for sequencing errors and artifacts |
By delineating the nuanced attributes of Sanger sequencing and NGS, researchers can navigate the intricacies of sequencing methodologies with informed precision, aligning their choice with the exigencies of the investigative or clinical domain at hand.
Advantages of NGS Over Sanger Sequencing
Amplified Sensitivity and Discovery Capacity
A pivotal advantage distinguishing NGS from Sanger Sequencing lies in its amplified sensitivity and capacity for discovery. NGS methodologies, characterized by their massively parallel sequencing paradigm, excel in discerning rare or low-frequency genetic variants with unparalleled depth. This heightened sensitivity assumes paramount significance in applications necessitating the revelation of novel mutations or infrequent genetic markers. Notably, in the realm of cancer research, where subtle mutations can precipitate disease progression, NGS's heightened sensitivity empowers researchers to delineate the complete gamut of genetic alterations harbored within tumor genomes. Such sensitivity augments initiatives in precision medicine by facilitating the pinpointing of actionable genetic targets, thereby fostering personalized treatment strategies.
Rapid Turnaround Time and Scalability
NGS presents a conspicuous advantage in terms of turnaround time and scalability when juxtaposed with Sanger Sequencing. By virtue of its capability to concurrently sequence millions of DNA fragments in a single operation, NGS platforms markedly truncate the duration requisite for generating comprehensive genomic data. This heightened throughput assumes particular significance in contexts characterized by high sample volumes, such as expansive population studies or clinical diagnostic facilities processing myriad samples on a daily basis. Moreover, the scalability inherent to NGS empowers researchers to expand their sequencing endeavors seamlessly, accommodating projects spanning a spectrum of magnitudes without compromising efficiency or cost-effectiveness. Consequently, NGS has emerged as the preferred modality for endeavors mandating expeditious data acquisition and analysis across diverse genomic terrains.
When to Use NGS vs. Sanger Sequencing
Sanger Sequencing: Focused Analysis and Single-Gene Investigations
Sanger Sequencing retains its pivotal role as a valuable tool for focused analysis and single-gene studies, particularly in contexts where precision and reliability take precedence. Benefitting from its meticulously established protocols and commendable accuracy rates, Sanger sequencing shines in scenarios necessitating the sequencing of individual genes or concise DNA fragments. Moreover, Sanger sequencing proves economically viable for endeavors entailing a restricted number of targets or samples, rendering it well-suited for modest-scale studies or diagnostic assays honing in on specific genetic loci. Furthermore, the familiarity and user-friendly nature of Sanger sequencing render it accessible to laboratories of varying expertise levels, thereby ensuring steadfast outcomes across diverse research environments.
NGS: High-Throughput Genomics and Comprehensive Analysis
NGS emerges as the favored approach for high-throughput genomics and exhaustive genomic scrutiny, presenting unmatched throughput and breadth of coverage. Its capacity to simultaneously scrutinize hundreds to thousands of genes empowers researchers to dissect intricate biological systems and unveil novel genetic variants spanning entire genomes or targeted regions. Furthermore, NGS's heightened sensitivity and scalability render it well-suited for applications necessitating the analysis of diverse sample types or extensive cohorts, such as population-scale genomics or clinical trials. By harnessing the capabilities of NGS, researchers stand poised to unearth fresh insights into the genetic foundations of disease, thereby paving the path for advancements in diagnostics, therapeutics, and personalized medicine.
Both Sanger Sequencing and NGS play pivotal roles in the field of genomics, offering distinct advantages and applications. While Sanger sequencing remains a reliable choice for targeted analysis and single-gene studies, NGS emerges as a transformative technology, empowering high-throughput genomics and comprehensive genomic analysis.
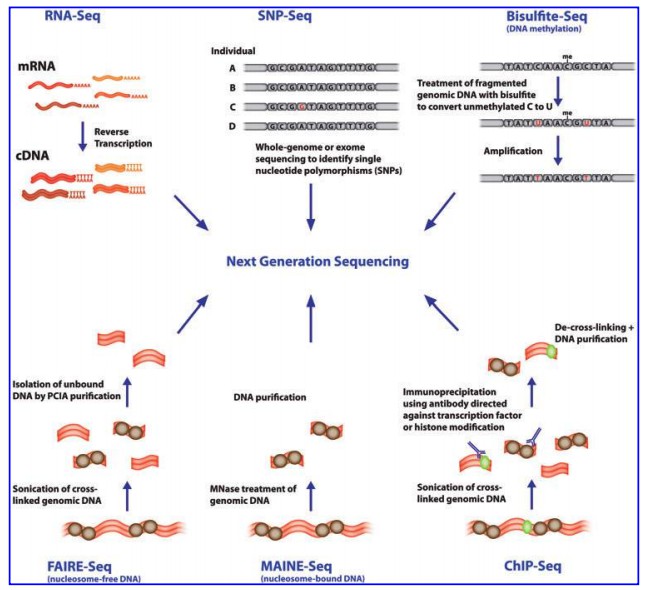 Applications of NGS (Evelien M. Bunnik et al,. 2012)
Applications of NGS (Evelien M. Bunnik et al,. 2012)
Illumina vs PacBio Sequencing
Illumina sequencing, a cornerstone of NGS technology, has catalyzed a paradigm shift in genomic research owing to its high-throughput capabilities and precision. Its utility traverses a broad spectrum, encompassing whole-genome sequencing to targeted amplicon analysis. Illumina's widespread adoption is underpinned by its resilience, accuracy, and cost-effectiveness, rendering it the favored option for large-scale genomic investigations, population genetics inquiries, and clinical diagnostics.
PacBio sequencing, colloquially known as Single-Molecule Real-Time (SMRT) sequencing, introduces a distinctive dimension to genomic analysis through its capacity to generate long reads. Diverging from Illumina, PacBio sequencing obviates the necessity for template amplification, mitigating biases and errors commonly associated with PCR. By monitoring the real-time incorporation of nucleotides, PacBio sequencing furnishes unprecedented insights into intricate genomic landscapes, elucidating structural variations, repetitive elements, and epigenetic modifications.
You may interested in
Learn More
PacBio vs Nanopore Sequencing
PacBio sequencing distinguishes itself by its capacity to generate extensive reads, providing invaluable insights into structural variations and genomic architecture. By observing the sequencing process in real-time, PacBio yields reads spanning thousands of base pairs, facilitating the accurate characterization of intricate genomic regions. This capability renders PacBio sequencing indispensable for diverse applications including de novo genome assembly, haplotype phasing, and full-length transcriptome analysis.
Nanopore sequencing emerges as a standout due to its portability and real-time analytical capabilities, offering a versatile tool for field research and point-of-care diagnostics. Through the traversal of DNA strands across nanopores embedded within lipid membranes, Nanopore sequencing discerns alterations in electrical currents, enabling direct determination of nucleotide sequences. This technology obviates the necessity for elaborate laboratory setups, facilitating swift and on-site genomic analyses. The applications of Nanopore sequencing span infectious disease surveillance, environmental monitoring, and personalized genomics, underscoring its broad utility in diverse domains.
You may interested in
Learn More
Nanopore vs Illumina Sequencing
Nanopore sequencing's portability and real-time analytical capabilities empower researchers to undertake genomic investigations in remote or resource-constrained settings. By harnessing handheld devices like the MinION, researchers can execute sequencing experiments beyond conventional laboratory confines, ushering in novel avenues for field research and point-of-care diagnostics. Nanopore's proficiency in directly sequencing long reads from DNA or RNA molecules confers advantages in diverse realms including de novo assembly, transcriptomics, and metagenomics.
Illumina sequencing, distinguished by its unparalleled throughput and precision, stands as the benchmark for large-scale genomic inquiries and clinical diagnostics. By engendering millions of short reads in parallel, Illumina sequencing ensures exhaustive coverage of the genome, facilitating variant discovery, gene expression profiling, and epigenetic investigations. Its robustness and cost-effectiveness have rendered it indispensable in endeavors necessitating high-resolution genomic data, encompassing domains like cancer genomics, population genetics, and infectious disease research.
In summary, Sanger Sequencing and NGS technologies such as Illumina, PacBio, and Nanopore sequencing offer complementary strengths in genomic analysis. Leveraging CD Genomics' proficiency across these sequencing platforms affords researchers access to a comprehensive array of genomic solutions, empowering them to explore the intricacies of the genome and embark on novel frontiers in biological research and clinical diagnostics.
References:
- Bunnik EM, Le Roch KG. An Introduction to Functional Genomics and Systems Biology. Adv Wound Care (New Rochelle). 2013
- Andrew D. Young, Jessica P. Gillung Phylogenomics — principles, opportunities and pitfalls of big-data phylogenetics. Systematic Entomology 2019


 Sample Submission Guidelines
Sample Submission Guidelines
