Illumina Library Preparation
Library construction involves preparing target RNA or DNA for compatibility with sequencing instruments. Initially, DNA or RNA is extracted from tissues or cells and fragmented using mechanical or enzymatic methods. In the case of RNA samples, reverse transcription is performed to convert RNA into cDNA, followed by ligation of DNA fragments. Subsequently, the library undergoes amplification, typically via PCR, to amplify the desired target fragments before it is ready for sequencing.
For Illumina library preparation, the process often includes specific steps tailored to Illumina sequencing platforms. This may involve end repair and the addition of dA bases to facilitate subsequent adapter ligation. Illumina library construction methods also incorporate unique adapter sequences compatible with Illumina sequencers. The libraries are then subjected to PCR amplification to enrich target fragments. Ultimately, Illumina libraries are prepared and optimized to yield high-quality sequencing data on Illumina platforms.
At the core of library construction lies the process of appending junctions to the ends of the fragments under examination. Presently, DNA library construction methods can be classified into five categories based on the different approaches to junction connection.
CD Genomics offers Illumina sequencing platforms that facilitate the robust analysis of DNA/genomes. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
Workflow of Library Preparation for Illumina Platforms
- End Repair and dA Tailing
After the random fragmentation of DNA in the preceding step, the generated DNA fragments may possess uneven ends. Thus, this step focuses on modifying the ends of these DNA fragments. It involves flattening the ends and appending a distinctive A base at the 3' end, while phosphorylating the 5' end. The primary objective of adding the A base is to create sticky ends, facilitating the attachment of junction primers in the subsequent step.
- Adaptor Ligation
An adaptor serves as a short sequence of bases, bridging the DNA fragment under examination to the Flowcell. T4 DNA ligase is commonly employed to mend single-stranded incisions in double-stranded DNA, facilitating the reconnection of adjacent nucleotides. In the case of ligation, a DNA fragment with a "T" sticky end can be linked to another DNA fragment with an "A" sticky end, forming a complete double strand.
Once the TA ligation occurs, the phosphodiester bond between the adaptor and the DNA fragment is established. Typically, the ligation reaction takes place in a buffer containing high-concentration salt particles and PEG. The high-concentration salt particles neutralize the negative charge of the phosphate backbone, facilitating nucleic acid aggregation, while PEG seizes water molecules from the nucleic acid backbone, promoting nucleic acid molecule aggregation. Additionally, PEG increases the solution's viscosity, aiding the settling of purification beads. However, magnetic beads may pose challenges in settling. Following ligation, the product requires purification to prevent interference with the subsequent PCR amplification step.
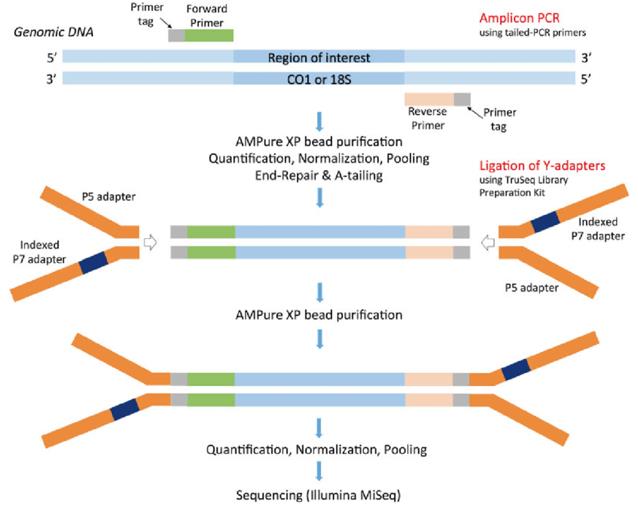 Scheme for Illumina MiSeq multiplex library preparation using the tailed PCR primers and ligation of single-indexed Y-adapters. (Bourlat et al., 2016)
Scheme for Illumina MiSeq multiplex library preparation using the tailed PCR primers and ligation of single-indexed Y-adapters. (Bourlat et al., 2016)
- Library Amplification
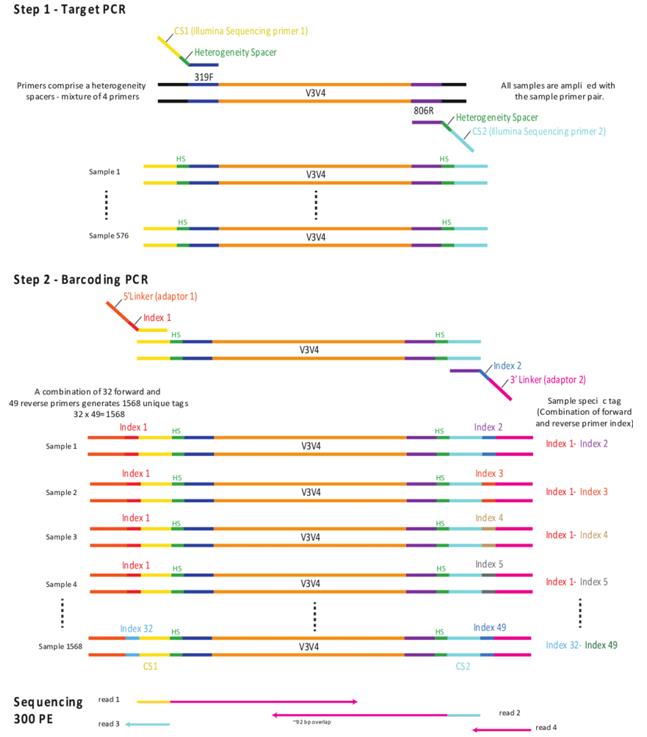 Illumina amplicon library preparation through 2-Step PCR amplification. (Holm et al., 2018)
Illumina amplicon library preparation through 2-Step PCR amplification. (Holm et al., 2018)
Comparing Illumina Library Preparation Methods: TruSeq DNA vs. Nextera
Illumina offers two primary methods for library preparation: the TruSeq DNA (standard library construction) and Nextera library construction methods (transposase method for library construction), each catering to distinct needs and preferences.
The TruSeq DNA method is recognized for its complexity, yet it offers several advantages. With a library starting volume requirement of 500ng, it accommodates lower-quality DNA samples, presenting a cost-effective solution with high genome coverage. Moreover, its automation capabilities streamline the process, making it suitable for general genome libraries.
On the other hand, the Nextera library construction method is prized for its simplicity. Although it demands higher quality DNA, it boasts advantages such as faster operation and flexibility. With a starting volume requirement as low as 50ng, or even 1ng, it is particularly well-suited for samples with limited volume. However, it comes at a higher cost and may result in lower genome coverage compared to the TruSeq method.
Illumina's Standard Library Construction
Illumina's standard library construction predominantly involves mechanical interruption for fragment sizing, with the exception of amplicon construction. The library assembly procedure unfolds as follows:
- DNA fragmentation via ultrasonic interruption.
- End repair, followed by addition of "A".
- Junction joining.
- Fragment screening.
- PCR enrichment of target fragments, followed by quality control.
- Sequencing.
The Transposase Method for Library Construction
The transposase method for library construction employs transposase to fragment the genome, and the process unfolds as follows:
Transposase fragments the genomic DNA, simultaneously integrating the required adaptors directly onto the ends of the fragments, thereby shortening library construction time and reducing sample requirement. This method is particularly suitable for applications with limited sample availability, such as tumor biopsy, degraded DNA, or purified DNA.
- Attachment of junctions to transposase-interrupted fragments.
- Purification of library DNA fragments (transposase removal).
- Several cycles of PCR to incorporate P5, P7 junctions, and indexes.
- Fragment screening for quality control.
- Sequencing.
Illumina DNA PCR-Free Library Preparation
In the Illumina DNA PCR-Free Prep Tagmentation process, the following steps are involved:
The product name has transitioned from Nextera DNA Flex to Illumina DNA Prep, reflecting its reliance on the on-bead tagmentation method.
This innovative method combines DNA extraction, fragmentation, library preparation, and library normalization into a single streamlined process. Initially, BLT (Bead-Linked Transposomes) enzymatically fragment the genomic DNA (gDNA) while simultaneously attaching Illumina's sequencing primers. Subsequently, the fragmented gDNA undergoes PCR amplification, incorporating sequence tags and junctions. Afterward, the sequenced fragments undergo purification, and the library is prepared for sequencing.
Other NGS Library Construction Methods
NGS library construction can be categorized into five types based on the different approaches to joining target fragments.
- TA Cloning Ligation Method: This widely used method is applicable to most samples and represents the mainstream commercial library construction approach.
- Swift Method: Similar to the TA cloning ligation method, the Swift method involves slightly more cumbersome operations. Notably, the connection of P5 and P7 adaptors occurs in two steps.
- Transposase Method: This method completes library construction in just 1 hour and 40 minutes, significantly reducing manpower requirements. However, it is only suitable for constructing cDNA and whole genome libraries. Additionally, the transposase enzyme exhibits preferences that may impact sequencing quality.
- PCR Amplicon Library Construction: This approach, known as capture library construction, is particularly useful for researching targeted genes within clinical contexts.
- Flat-End Junction Library: Specifically designed for the Ion Torrent platform, this method is limited in application due to the relatively low market share of Ion Torrent. Therefore, its usage is relatively restricted.
References:
- Holm, Johanna B., et al. "Ultra-high throughput multiplexing and sequencing of> 500 bp amplicon regions on the Illumina HiSeq 2500 platform." bioRxiv (2018): 417618.
- Bourlat, Sarah J., et al. "Preparation of amplicon libraries for metabarcoding of marine eukaryotes using Illumina MiSeq: the dual-PCR method." Marine genomics: Methods and protocols (2016): 197-207.


 Sample Submission Guidelines
Sample Submission Guidelines
