What is Next Generation Sequencing (NGS)?
History of Next-generation Sequencing
Next Generation Sequencing (NGS) is a revolutionary genetic analysis technique that involves fragmenting the genetic material (DNA or RNA) and attaching oligonucleotides with known sequences through a process called adapter ligation. This allows the resulting fragments to interact with the selected sequencing platform. Subsequently, the bases within each fragment are identified based on their emission signals.
The key distinction between Sanger sequencing and NGS lies in the sequencing scale; NGS can simultaneously process millions of reactions, providing exceptional throughput, heightened sensitivity, rapid results, and cost-effectiveness. It is now feasible to complete numerous genome sequencing projects in a matter of hours, a task that would have taken much longer using traditional Sanger sequencing methods.
NGS technology primarily follows two main approaches: short-read and long-read sequencing, each offering its own unique advantages and limitations. The driving force behind extensive investment in NGS development is its versatile applicability in both clinical and research contexts.
For long-read sequencing, please refer to the articles Nanopore Sequencing: Principles, Platforms and Advantages and Overview of PacBio SMRT sequencing: principles, workflow, and applications for more information.
Next-generation Sequencing Platforms and Technologies
Next-generation sequencing methods are well-established and share common characteristics, yet they can be categorized based on their underlying detection chemistry. These categories include sequencing by ligation and sequencing by synthesis (SBS).
Please refer to CD Genomics Sequencing Platforms.
The most prevalent SBS method is reversible terminator sequencing, which employs "bridge amplification." DNA fragments attach to oligonucleotides on a flow cell in this process, forming a bridge from one side of the sequence to the other. This bridge is then amplified, and fluorescently labeled nucleotides are detected using direct imaging.
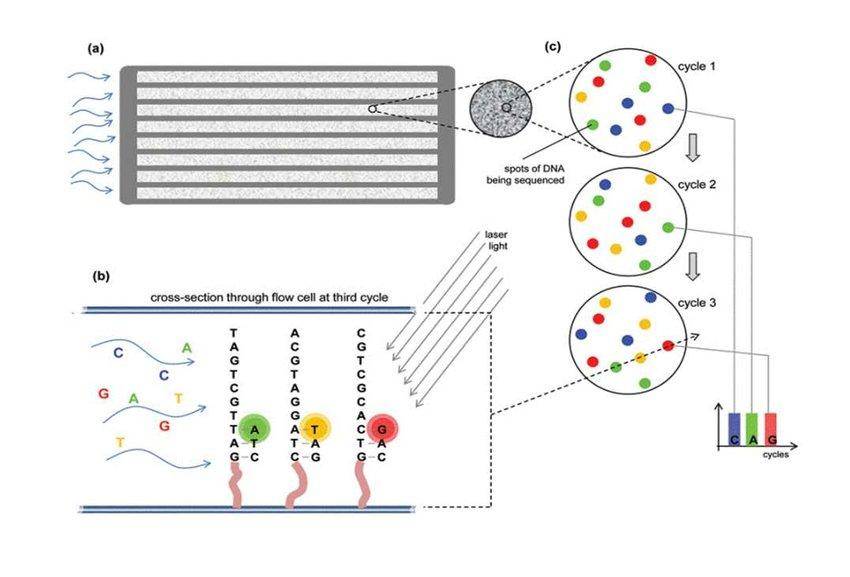 Principle of Illumina Sequencing (Sequencing by Synthesis). (Untergasser et al., 2019)
Principle of Illumina Sequencing (Sequencing by Synthesis). (Untergasser et al., 2019)
In contrast to SBS, sequencing by ligation doesn't require DNA polymerase to generate a second strand. Instead, fluorescence signals are used to identify the target sequence, capitalizing on the sensitivity of DNA ligase to detect base pair mismatches.
NGS technologies typically offer multiple advantages over alternative sequencing methods, allowing for rapid, sensitive, and cost-effective sequencing reads. Nonetheless, there are drawbacks, such as challenges in interpreting homopolymers and polymerase errors due to incorrect dNTP incorporation, which can result in sequencing errors.
A major limitation of all NGS technologies is the requirement for PCR amplification before sequencing, which introduces biases during library preparation (related to sequence GC content, fragment length, and pseudo diversity) and during analysis (resulting in base errors and favoring specific sequences over others).
PacBio SMRT Sequencing
PacBio Single Molecule Real-Time (SMRT) Sequencing leverages the concept of sequencing while synthesizing, using the SMRT chip as the sequencing medium. This chip contains numerous tiny pores, each housing a DNA polymerase molecule. During the base pairing process, distinct nucleotide bases are incorporated, emitting unique wavelengths and peak values of light. These emitted signals serve to determine the type of base incorporated. Remarkably, SMRT sequencing operates at an astonishing pace, processing several deoxyribonucleoside triphosphates (dNTP) per second.
Oxford Nanopore Sequencing
Oxford Nanopore Sequencing stands out from previous sequencing technologies as it relies on electrical signals rather than optical ones. A pivotal component of this technology is a specially designed nanopore, capable of accommodating only a single DNA molecule, featuring covalently attached molecular junctions within. As DNA bases traverse the nanopore, they induce transient changes in electrical charge, affecting the current strength passing through the nanopore. Each base type results in a distinct alteration in current magnitude, which is sensitively detected by electronics, enabling the identification of the transiting bases.
Steps in Next-generation Sequencing Library Preparation
(1) Sample Preparation (Pre-treatment)
Nucleic acids (DNA or RNA) are extracted from chosen samples (e.g., blood, sputum, bone marrow). The extracted samples undergo quality control (QC) assessment using standard methods such as spectrophotometry, fluorimetry, or gel electrophoresis. If RNA is employed, it may require reverse transcription to generate cDNA, although some library preparation kits may incorporate this step.
Please refer to our Sample Submission Guidelines for more information.
(2) Optimization and Enhancement of NGS Libraries
cDNA or DNA is typically randomly fragmented using enzymatic treatment or sonication. The optimal fragment length depends on the sequencing platform being used. Optimization may entail running a small subset of fragmented samples on an electrophoresis gel. These fragments are subsequently end-repaired and linked to shorter generic DNA fragments known as adapters. These adapters possess a defined length and known oligo sequences compatible with the chosen sequencing platform, enabling recognition during multiplex sequencing. Multiplex sequencing, employing the respective adapter sequences for each sample, permits the simultaneous sequencing of numerous libraries in a single run. The pool of DNA fragments with adapters is referred to as the sequencing library.
Please refer to our article Quality Control in the NGS Library Preparation Workflow for more information.
Size selection can be achieved through gel electrophoresis or the use of magnetic beads to eliminate excessively short or long fragments that may not perform optimally on the selected sequencing platform and protocol. PCR is then employed to amplify/enrich the library. In techniques involving emulsion PCR, each fragment is tethered to an individual emulsion bead, which forms the basis of the sequencing cluster. A "clean-up" step, often utilizing magnetic beads, is performed post-amplification to eliminate undesired fragments and enhance sequencing efficiency.
The final library can be subjected to quality control using quantitative PCR (qPCR) to confirm both the quality and quantity of DNA. This step also facilitates the preparation of samples with the appropriate concentration for sequencing.
(3) Sequencing
Depending on the chosen platform and chemistry, clonal amplification of library fragments can occur either before loading the sequencer (PCR) or on the sequencer itself (bridge PCR). Sequences are then detected and reported based on the selected platform.
(4) Data Analysis
Data files generated are analyzed in accordance with the specific workflow employed. Analysis methods are highly dependent on the study's objectives.
While paired-end and paired-pair sequencing can reduce the number of samples analyzed in a single run, they offer distinct advantages in downstream data analysis, especially for de novo assembly. These technologies combine sequencing reads obtained from both ends of a fragment (paired-end) or those separated by interstitial DNA regions (paired pairs).
How to Choose an Appropriate Library Preparation and Sequencing Platform?
(a) Articulating the research question
(b) Selecting the sample type
(c) Deciding between short-read or long-read sequencing
(d) Determining if DNA or RNA sequencing is required (genome or transcriptome analysis)
(e) Defining the scope, whether it involves the entire genome or specific regions
(f) Establishing the necessary read depth (coverage) tailored to the experiment
(g) Considering the extraction method
(h) Evaluating sample concentration
(i) Selecting between single-ended, paired-end, or paired-pair reads
(j) Specifying the required read length
(k) Exploring the feasibility of multiplexing samples
(l) Assessing the bioinformatics tools needed, which varies depending on the experiment. The entire sequence analysis process can be adapted based on the sample and the biological question at hand.
Next-generation Sequencing Data Analysis
Every Next Generation Sequencing (NGS) technology generates an extensive volume of output data. The foundational sequence analysis workflow is centralized and encompasses several key steps: raw read quality control, data preprocessing and alignment, followed by post-processing, variant annotation, variant calling, and visualization.
The initial step in this workflow involves evaluating the quality of raw sequencing data, a crucial prerequisite for all downstream analyses. This assessment provides essential insights into the overall data, including the quantity and length of reads, the presence of contaminating sequences, and any reads with inadequate coverage.
Please refer to our service Bioinformatic Service for Next Generation Sequencing for more information.
Challenges in Next-generation Sequencing
NGS has revolutionized our ability to explore and study genomes. In clinical settings, NGS plays a crucial role in diagnosing a wide range of diseases by detecting germline or somatic mutations. The increasing adoption of NGS in clinical practice is justified by its ability to effectively pair advanced technology with decreasing costs.
Moreover, NGS is an indispensable tool in the field of metagenomics research, enabling the diagnosis, surveillance, and management of infectious diseases. In 2020, NGS methods played a pivotal role in characterizing the SARS-CoV-2 genome and continue to be essential for monitoring the ongoing COVID-19 pandemic.
However, the intricacies of NGS sample processing reveal significant challenges in the management, analysis, and storage of data. One of the primary challenges is the substantial computational resources required for tasks such as data assembly, annotation, and analysis.
Additionally, the sheer volume of data produced by NGS poses a substantial hurdle. Data centers are grappling with high storage demands and struggling to keep pace with the increasing data load, which raises concerns about the risk of permanent data loss. Continuous efforts are underway to enhance efficiency, reduce sequencing errors, maximize reproducibility, and ensure robust data management in order to address these challenges.
Reference:
- Untergasser, Gerold & Bucher, Philipp & Dresch, Philipp. (2019). Metagenomics Profiling of Tumours Using 16S-rRNA Amplicon Based Next Generation Sequencing.


 Sample Submission Guidelines
Sample Submission Guidelines
