Quality Control in NGS Library Preparation Workflow
Next-generation sequencing (NGS) has revolutionized genomics, enabling researchers to delve into the intricacies of DNA and RNA at an unprecedented scale. However, the success of NGS heavily relies on the quality of the prepared libraries. To optimize time and resources, efficient quality control (QC) checks are indispensable at crucial steps in the library preparation workflow. Implementing QC measures before and after various stages of the process ensures high-quality samples, minimizes costly errors, and enhances the overall success of NGS experiments.
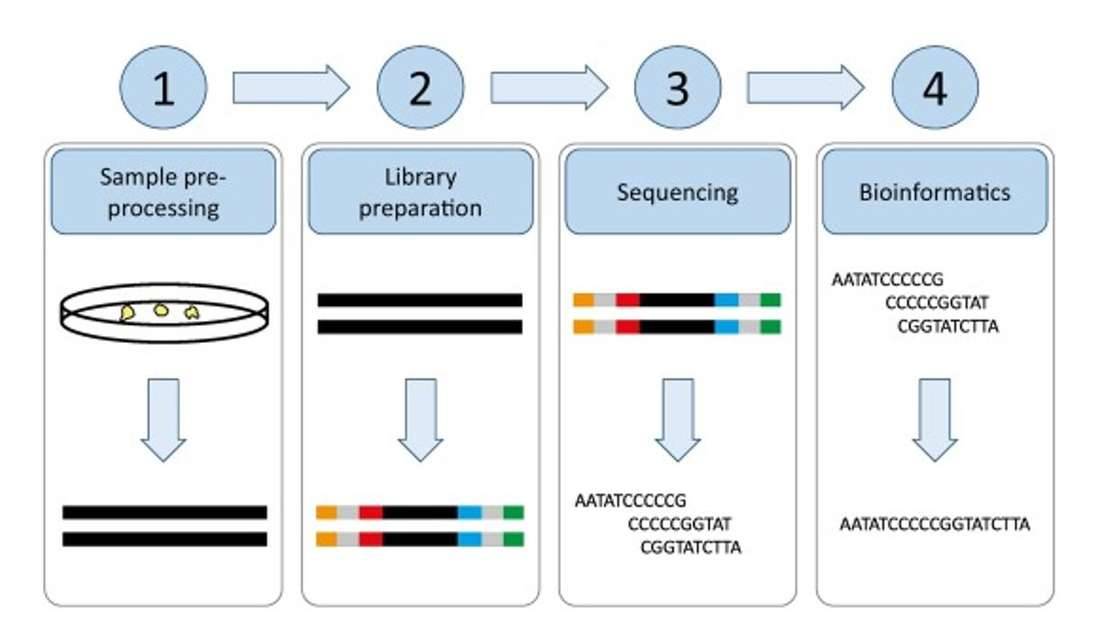 Next generation sequencing. (Hess et al., 2020)
Next generation sequencing. (Hess et al., 2020)
The Importance of Sample Quality in NGS Library Preparation
NGS library preparation involves several critical steps, including sample fragmentation, ligation of sequencing adapters, and library amplification. The quality of the starting samples significantly impacts the success of these steps and, consequently, the sequencing results. Poor quality or insufficiently concentrated samples can lead to biased or unreliable data, potentially wasting valuable resources and delaying research outcomes. By performing QC checks throughout the process, researchers can identify potential issues early on and optimize their protocols for optimal results.
Please refer to our Sample Submission Guidelines for more details.
Starting Material QC in NGS Library Preparation
The success of NGS heavily relies on the quality of the starting materials used in the library preparation process. Adequate quality control (QC) of the starting materials, whether DNA or RNA, is the first and crucial step in preparing high-quality libraries and obtaining successful sequencing results.
The starting material, such as genomic DNA or total RNA, serves as the foundation for NGS library construction. It is essential to assess the quality and quantity of these samples before initiating library preparation. High-quality starting materials significantly impact the downstream processes, ensuring accurate and representative sequencing data. Conversely, degraded or low-quality samples can lead to biased results, loss of valuable sequencing material, and compromised library complexity.
Critical QC Parameters for Starting Material:
- Quantity: Accurate quantification of starting material is essential to determine the appropriate amount needed for library preparation. NGS library preparation kits often recommend specific starting concentrations and amounts to optimize sequencing results. Precise quantification ensures that the library is not under or over-sequenced, leading to optimal coverage and data generation.
- Purity: The purity of the starting material is evaluated by measuring the absorbance ratios at specific wavelengths, typically A260/A280 and A260/A230. An A260/A280 ratio close to 1.8 and A260/A230 ratio around 2.0 indicates minimal contamination from protein, organic compounds, or salts. Contaminants can interfere with enzymatic reactions during library preparation, potentially affecting downstream analysis.
- Integrity: The integrity of the nucleic acid sample is crucial for successful library preparation. For RNA samples, RNA integrity number (RIN) or RNA quality number (RQN) can be determined using techniques like capillary electrophoresis. High RIN/RQN scores indicate intact RNA molecules, while degraded RNA may result in fragmented and biased library construction.
Reproducibility: Ensuring that the starting material is reproducible is essential, especially in multi-sample studies. Replicates of the same sample should yield consistent QC values to minimize technical variability during library preparation.
QC Checkpoints in NGS Library Preparation
To ensure accurate and reliable sequencing data, it is crucial to implement quality control (QC) checkpoints at various stages throughout the NGS library preparation process. This article outlines the significance of QC at different points in the workflow to guarantee the creation of high-quality libraries for successful sequencing.
QC Checkpoints in NGS Library Preparation:
- Sample QC
The initial QC step involves assessing the quality and quantity of the starting nucleic acid samples (DNA or RNA) before library preparation. Accurate quantification, purity assessment, and integrity checks of the samples are essential to avoid introducing bias and ensure optimal representation of the original material in the library.
- Fragmentation QC
After quantification, the samples undergo fragmentation, breaking them into smaller DNA or RNA fragments. QC at this stage confirms that the fragmentation process was successful and that the fragment size distribution is within the desired range for the specific experiment.
- Splice Ligation QC
Following fragmentation, the sequencing adapters are ligated to the fragmented DNA or RNA. QC checks are performed to validate the efficiency of the ligation process and ensure that adapters are successfully incorporated, facilitating proper attachment to the sequencing platform.
- Amplification QC
PCR amplification is a crucial step to enrich the library with sequencing-ready material. QC checks at this stage verify that the amplification was successful without over-amplification or under-amplification, which can introduce biases and affect the library complexity.
- Adapter Dimer Detection
Throughout the library preparation process, it is essential to monitor and detect the presence of adapter dimers. These artifacts can form when excess adapters ligate together, reducing the representation of the original material in the library. QC checks are performed to minimize adapter dimer content, ensuring a high-quality library.
- Library Pooling QC
In multi-sample studies, libraries are often pooled together for multiplexed sequencing. Pooling QC involves quantifying and normalizing the concentration of individual libraries to ensure equal representation of each sample, maximizing sequencing efficiency and data yield.
QC for the Final NGS Library
The final step in the NGS library preparation process is PCR amplification, a critical step that generates sufficient copies of the library for subsequent sequencing. During this stage, ligation of sequencing adapters to the fragmented DNA or RNA is confirmed, and PCR amplification is employed to enrich the target DNA fragments. Quality control (QC) of the final NGS library is essential to ensure the success of the sequencing run, identify potential issues, and obtain reliable and high-quality sequencing data.
Importance of QC for the Final NGS Library:
The final NGS library represents the DNA or RNA fragments that are ready for sequencing. Assessing the quality of the library at this stage is crucial because it determines the success of the subsequent sequencing run. QC of the final library helps to:
- Verify Successful Library Preparation: Confirm that the ligation of sequencing adapters and other library preparation steps were successful, ensuring that the library is suitable for sequencing.
- Detect Adapter Dimers: Identify the presence of excess primers or adapter dimers, which can interfere with the sequencing process and lead to the misrepresentation of the original sample.
- Assess Amplification Efficiency: Determine whether the library has been adequately amplified. Over-amplification can result in increased duplicates and biases, while under-amplification can lead to insufficient representation and reduced sequencing yield.
- Optimize Library Pooling: Quantify the concentration and molar concentration of the library to facilitate proper pooling of multiple libraries for multiplexed sequencing, ensuring equal representation of each sample.
QC Methods for the Final NGS Library:
- Electrophoresis: Automated electrophoresis platforms, such as Bioanalyzer or TapeStation, are commonly used for fragment size analysis of the final library. The presence of a clean library dispersion peak confirms successful library preparation, while the presence of additional peaks indicates the presence of adapter dimers or other contaminants.
- Quantification: Fluorometric methods like Qubit or qPCR can accurately quantify the concentration and molar concentration of the library, enabling precise pooling of libraries for sequencing.
Library Complexity Analysis: Techniques like molecular barcodes or other indexing strategies can be employed to assess library complexity and detect PCR duplicates, which are essential for detecting and correcting potential biases introduced during PCR amplification.
Reference:
- Hess, Jacob Friedrich, et al. "Library preparation for next generation sequencing: A review of automation strategies." Biotechnology advances 41 (2020): 107537.


 Sample Submission Guidelines
Sample Submission Guidelines
