Overview of PacBio SMRT sequencing: principles, workflow, and applications
Overview of PacBio sequencing
Pacific Biosciences (PacBio) sequencing embodies a vital third-generation sequencing method, in the technical parlance, dubbed "Single Molecule Real-Time (SMRT) DNA Sequencing". This direct approach circumvents the requirement for DNA amplification, thereby facilitating prompt sequence reading. Contrasting this with conventional sequencing procedures which necessitate fragmenting DNA into smaller portions for sequencing, PacBio sequencing, on the other hand, performs real-time reading of the entire DNA molecule as it passes through a specialized sequencing apparatus.
Since its inception in 2005, PacBio has been a pioneer in the field of genomics with the advent of its unique Single Molecule Real Time (SMRT) sequencing technology. By harnessing the power of Zero Mode Waveguide (ZMW) arrays, SMRT technology allows for the real-time sequencing of DNA molecules of up to tens of thousands of base pairs in length.
In 2010, PacBio unveiled its premier SMRT sequencing system, the PacBio RS, which boasted advantages such as high throughput, long reads, and precision. By 2015, after considerable improvements in reagent and analysis software, PacBio's RSII platform began to gain traction in the field of genomics research. A constant stream of papers utilizing this technology was published in renowned journals such as Nature and Science. Specifically, several human genome research papers, tackling everything from genome enhancement to the development of assembly algorithms, have appeared in quick succession.
In recent years, PacBio has continuously refined its sequencing technology, leading to the launch of the Sequel II and Sequel IIe systems. These second-generation platforms significantly augment sequencing efficiency and accuracy, thanks to upgraded hardware and software that support on-instrument CCS processing. This allows customers to directly utilize HiFi reads for subsequent analyses. These enhancements minimize workflow complexity, expedite the delivery of results, and reduce the overall system cost, contributing to the further democratization of genomics research.
Service you may intersted in
Principle of PacBio SMRT sequencing
The utilization of PacBio sequencing embraces a methodology characterized by concurrent synthesis and sequencing, employing SMRT chip as the sequencing vector. This technique operates on the principle of DNA polymerase activity. Fundamentally, DNA polymerase binds to the template DNA, where each of the four nucleotide bases (dNTPs) is fluorescently labeled with distinct colors.
During the base-pairing phase, the incorporation of different nucleotides emits varying wavelengths of light, discernible by their respective peaks and wavelengths. Moreover, the DNA polymerase enzyme plays a pivotal role in achieving exceptionally long reads, a characteristic attributed to its enzymatic activity. The length of reads primarily correlates with the maintenance of enzyme activity, which in turn is influenced significantly by the damage inflicted by laser irradiation.
The construction of the third-generation PacBio library involves connecting the ends of the double-stranded DNA molecules (fragments of lengths ranging from 10-20kb or longer) with PacBio adaptors that possess hairpin structures. This results in the DNA molecule assuming a dumbbell-shaped structure known as the SMRTbell library. During sequencing, the polymerase emits a fluorescent signal under the influence of the fluorescent-tagged nucleotides. This signal is collected by a charge coupled device (CCD) camera. The procedure is iterated on the cyclic library, thereby enabling sequencing to be completed.
The PacBio SMRT sequencing utilizes the innovative Zero-Mode Waveguide (ZMW) technology to differentiate ideal fluorescence signals from the intense fluorescent backgrounds caused by freely floating nucleotides. The bonded DNA polymerase and the template DNA strand are affixed to the glass surface at the base of the ZMW. Lasers percolate through the base of the ZMW, but do not fully penetrate it due to the fact that the size of the ZMW is smaller than the wavelength of light. Thus, it permits selective excitation and recognition of light emitted from the nucleotides used for base extension.
Workflow of PacBio SMRT sequencing
The workflow of PacBio sequencing involves steps such as sample preparation, library construction, sequencing reaction, data analysis, and interpretation of results.
Sample preparation primarily involves the isolation of DNA from the samples to be tested. DNA samples can be sourced from bacteria, plants, animals, or human cells.
Library construction encompasses several steps: assessing the quality of genomic DNA (gDNA), shearing gDNA using a g-TUBE (Covaris); selecting size and adjusting concentration; repairing DNA damage and DNA fragment ends; purifying DNA; undertaking blunt-end ligation using hairpin adaptors; and purifying the template for submission to the sequencer.
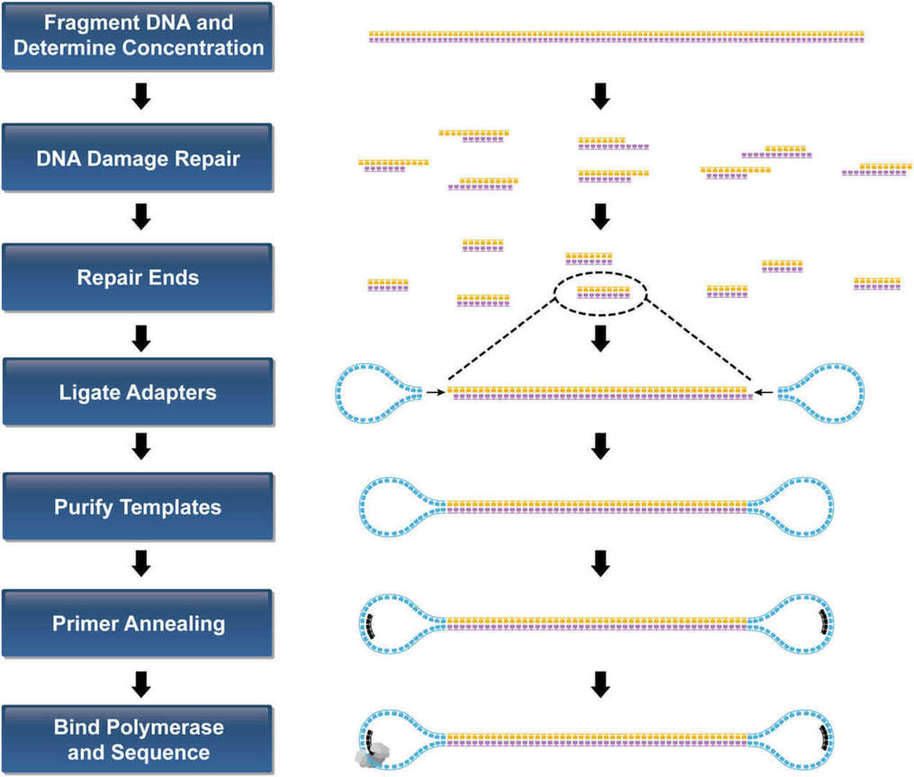 Figure 1. Template Preparation Workflow for PacBio RS II system.
Figure 1. Template Preparation Workflow for PacBio RS II system.
For the sequencing reaction, as illustrated in Figure 2, SMRTbell (shown in grey) diffuses into the Zero-Mode Waveguide (ZMW) and the adaptor binds with the polymerase fixed at the bottom. The four types of nucleotides are tagged with distinct fluorescent dyes (represented as red, yellow, green, and blue, corresponding to G, C, T, and A respectively), thus they have different emission spectra. The polymerase produces a pulse of light that identifies the base when it retains the nucleotide in the detection volume.
The reaction involves the fluorescently tagged nucleotide binding to the template at the polymerase active site. (1) The fluorescence output heightens corresponding with the incorporated base (here C depicted as yellow). (2) The labelled pyrophosphate-dye conjugate cleaves from the nucleotide and diffuses out of the ZMW, terminating the fluorescent pulse. (3) The polymerase advances to the next position. (4) The succeeding nucleotide binds to the template at the polymerase active site, initiating the subsequent fluorescent pulse, corresponding to base A in this context.
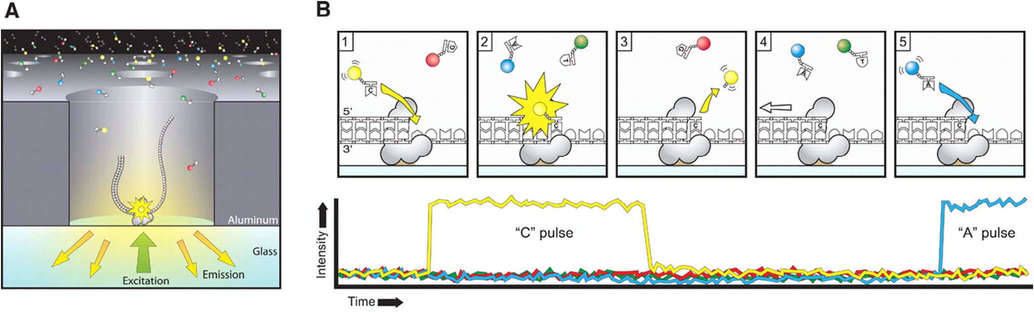 Figure 2. Sequencing via light pulses.
Figure 2. Sequencing via light pulses.
Bioinformatics analysis, such as de novo assembly, reference genome mapping, genomic annotation (prediction of pathogenic and susceptibility genes, non-coding RNA prediction, CRISPRs prediction), gene function annotation (COG/GO/KEGG), SNP/InDel identification, comparative genomics analysis, and evolutionary analysis and divergence time estimation, are viable steps.
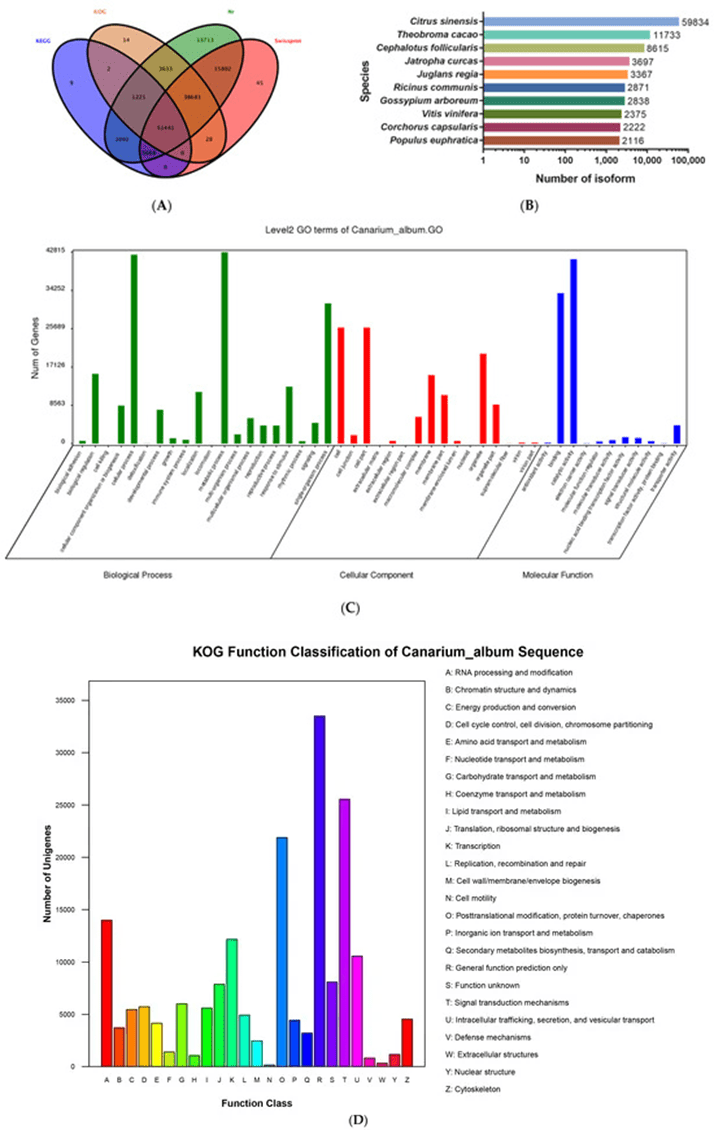 Figure 3. Full-length transcriptome sequencing and assembly of C. album by the SMRT method. (Ye et al., 2024)
Figure 3. Full-length transcriptome sequencing and assembly of C. album by the SMRT method. (Ye et al., 2024)
Advantages of PacBio Sequencing
PacBio sequencing has many excellent features as following:
Exceptionally long sequencing read length: The average sequencing read length can reach 8-15kb, with the longest sequences reaching up to 40-70kb.
High accuracy: For genome assembly and genomic variant detection, the accuracy of up to 99.999% can be achieved. By using a special sequencing mode, sequencing accuracy can reach 99% at the single-molecule level with a read length surpassing the classic Sanger sequencing method.
Extreme sensitivity: Can detect minor variants with a frequency of 0.1%.
Direct broad base modification detection: Aside from 5-methylcytosine modification detection, N6-methyladenine, N4-methylcytosine, DNA oxidation damage, and other base modifications can also be identified.
Minimum GC bias: Comfortable detection is made possible within areas of extreme high GC and extreme low GC, thereby ensuring uniform sequence coverage.
No PCR amplification bias: PCR amplification is not required for samples, thus avoiding uneven coverage and PCR redundancy.
Epigenetics: As there is no PCR amplification stage, base modifications can be directly detected during sequencing. The need for chemical modifications to detect base modifications is discarded owing to the measurement of polymerase kinetics changes during DNA base incorporation. This enables simultaneous capture of sequence and epigenetic information within a single experiment.
Differnences among PacBio, Illumina and Nanopore
The principle of Illumina sequencing relies on the accumulation of thousands of fluorescence signals to accurately sequence individual nucleotides, yielding relatively short reads for data processing, with read lengths up to 600bp and a minimum error rate of 1%. In addition to high accuracy, it offers large data output and relatively cheaper prices. However, Illumina sequencing, based on PCR, tends to exhibit GC bias and is limited by its shorter read lengths, making it unsuitable for assembling long repetitive sequences and restricting its broader applications, such as genome assembly and detection of long non-coding RNAs.
PacBio sequencing, also known as Single Molecule Real-Time (SMRT) sequencing, utilizes Zero-Mode Waveguide (ZMW) technology to confine individual DNA molecules and DNA polymerase within a ZMW pore, enabling sequencing while synthesizing, thereby easily obtaining read lengths of tens of kilobases with an average read quality of up to 99%. However, due to the independence of ZMW pores and real-time sequencing, the error rate of reads once reached 15%, posing significant challenges for subsequent data analysis. Recently, PacBio introduced the CCS mode, which drastically reduces the error rate by multiple sequencing of individual molecules. This sequencing method is particularly suitable for genome assembly, DNA methylation, RNA methylation, structural variation detection, and Long-reads transcriptome analysis.
Nanopore sequencing achieves long read lengths of up to several tens of kilobases by guiding DNA or RNA molecules through nanopores on a membrane, with records exceeding megabases (such as obtaining 2.3Mb sequencing data in the human genome). This sequencing method does not rely on PCR amplification, thus avoiding GC bias issues. Although the error types primarily consist of insertions and deletions, they are random, and error rates can be reduced by repeated sequencing. This makes nanopore sequencing advantageous in genome assembly, direct RNA sequencing, and somatic mutation detection.
In conclusion, each of these three sequencing methods has its own advantages and disadvantages, and the choice of sequencing strategy should be based on the specific requirements of the research.
Service you may intersted in
Applications of PacBio SMRT sequencing
PacBio SMRT sequencing has multifarious applications such as de novo whole-genome sequencing, optimization or mapping of genomic drafts, whole transcriptome sequencing, metagenomic sequencing, full-length 16S rRNA sequencing, organellar genome sequencing, whole genome resequencing & identification of rare variants, epigenetics and etc. These diverse applications render PacBio SMRT sequencing a formidable tool in the field of genomics research.
De novo assembly: PacBio's long read length significantly enhances the success rate of contig assembly, producing long contigs. It can effortlessly traverse repetitive and high GC sequences. In practice, researchers typically use PacBio sequences for contig assembly and then employ Illumina sequences for base correction.
Human leukocyte antigen (HLA) typing: Accurate HLA typing is critical in human organ transplantation. HLA encompasses a long fragment, and its haplotype plays a crucial role in successful typing and transplantation. Medical practitioners are now attempting to utilize PacBio sequences as a solution for accurate HLA typing.
Methylation research: PacBio can directly read a variety of base modifications, including methylation of adenine and cytosine, and hydroxymethylation of cytosine, thereby conferring PacBio a unique advantage in base modification research.
RNA alternative splicing research: The prerequisite for analyzing RNA alternative splicing involves a long-read sequence extending beyond the variable splicing site on both sides. Existing sequencing methodologies, due to their shorter read lengths, do not exhibit high sensitivity to RNA alternative splicing. Here, PacBio fills the void.
Detection of multiple repeat sequences: Some diseases ensue from the repetition of certain repeat sequences beyond normal range, such as the up to 750 CGG repeats in Fragile X syndrome. These sequences were challenging to fully determine via direct sequencing in the past. Now, scientists can directly sequence these regions using PacBio.
References:
- Ye Q, Zhang S, Xie Q, et al. De Novo Transcriptome Analysis by PacBio SMRT-Seq and Illumina RNA-Seq Provides New Insights into Polyphenol Biosynthesis in Chinese Olive Fruit. Horticulturae, 2024, 10(3): 293.
- Kong N., Ng W., Thao K., et al. Automation of pacbio smrtbell ngs library preparation for bacterial genome sequencing, Standards in Genomic Sciences, 2017, 12(1), 27.
- Ye W, Xu W, Xu N, et al. Comprehensive transcriptome characterization of Grus japonensis using PacBio SMRT and Illumina sequencing. Scientific Reports, 2021, 11(1): 23927.
- Cuber P, Chooneea D, Geeves C, et al. Comparing the accuracy and efficiency of third generation sequencing technologies, Oxford Nanopore Technologies, and Pacific Biosciences, for DNA barcode sequencing applications. Ecological Genetics and Genomics, 2023, 28: 100181.
- Chen Z, He X. Application of third-generation sequencing in cancer research. Medical Review, 2021, 1(2): 150-171.
- PacBio's website.


 Sample Submission Guidelines
Sample Submission Guidelines
