Sanger Sequencing: Introduction, Workflow, and Applications
What Is Sanger Sequencing?
Sanger sequencing, a DNA sequencing method, hinges on the selective incorporation of chain-terminating dideoxynucleotides during the in vitro replication of DNA, guided by the action of DNA polymerase. This technique was pioneered by Frederick Sanger and his team in 1977, hence the term "Sanger Sequence". Crucial to the Sanger sequencing method is the phenomenon of DNA polymerase reaction termination instigated by dideoxynucleotide triphosphates (ddNTPs)—a principle that has led to the alternative name for the technique, the "dideoxy termination sequencing". Evidencing its efficacy and reliability, this method has been the most widely employed sequencing technique for nearly four decades.
The Sanger sequencing method, underpinned by an accuracy rate of 99.99% in base identification, is widely regarded as the 'gold standard' for verifying DNA sequences. This includes those DNA sequences initially deciphered using next-generation sequencing techniques. The Human Genome Project, for instance, utilized Sanger sequencing for determining the sequences of relatively short fragments of the human DNA, each comprising 900 base pairs or less. These fragments served as the building blocks for constructing larger DNA sections, which, in turn, were assembled to construct entire chromosomes.
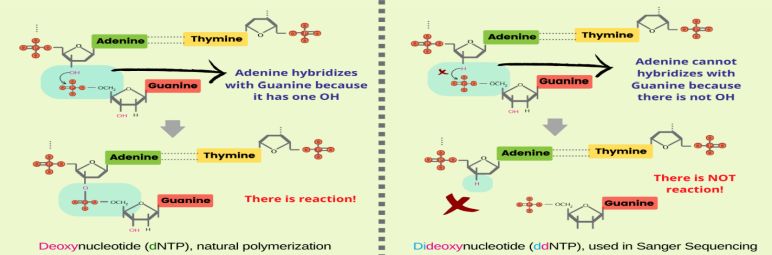 Figure 1. Chain termination reaction in Sanger sequencing. (Saini et al., 2023)
Figure 1. Chain termination reaction in Sanger sequencing. (Saini et al., 2023)
Services you may interested in
Sanger Sequencing Workflow
Sanger sequencing primarily entails the formation of a reaction system, amplification of the target segment, gel electrophoresis, and sequence reading. The process of Sanger sequencing consists of a set of four separate reactions, each encompassing: (1) the four deoxyribonucleotide triphosphates (dNTPs), critical for normal DNA synthesis; (2) the inclusion of four distinct didoxyribonucleotide triphosphates (ddNTPs) in each reaction system. Since ddNTPs are devoid of the requisite 3-OH group for elongation, the amplified oligonucleotides selectively terminate at G, A, T, or C. For effective positioning, these are marked with fluorescence or isotopes. (3) The target fragment, DNA polymerase, primers all constitute the reaction system.
Subsequently, the target fragment serves as the template under the catalysis of DNA polymerase, initiating DNA replication from the primers. When encountering ddNTPs, the reaction ceases. The ensuing gel electrophoresis aims to separate these terminated oligonucleotide fragments. Finally, the fragments are subject to electric current to induce band formation. Under the combined influences of electrical current and gel resistance, regular bands emerge longitudinally at equal intervals, representing varied sequence lengths, each corresponding to an A-T-G-C sequence.
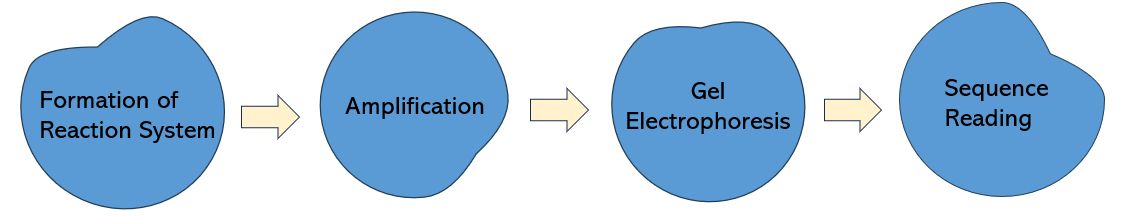 Figure 2. General workflow of Sanger sequencing
Figure 2. General workflow of Sanger sequencing
There are three main steps to Sanger sequencing which are the following:
DNA Sequence for Chain Termination PCR
The DNA sequence of interest is used as a template for a special type of PCR called chain-termination PCR. Chain-termination PCR works just like standard PCR, but with one major difference: the addition of modified nucleotides called dideoxyribonucleotides. In the extension step of standard PCR, DNA polymerase adds dNTPs to a growing DNA strand by catalyzing the formation of a phosphodiester bond between the free 3'-OH group of the last nucleotide and the 5'-phosphate of the next.
Size Separation by Gel Electrophoresis
In the second step, the chain-terminated oligonucleotides are separated by size via gel electrophoresis. In gel electrophoresis, DNA samples are loaded into one end of a gel matrix, and an electric current is applied; DNA is negatively charged, so the oligonucleotides will be pulled toward the positive electrode on the opposite side of the gel. Because all DNA fragments have the same charge per unit of mass, the speed at which the oligonucleotides move will be determined only by size. The smaller a fragment is, the less friction it will experience as it moves through the gel, and the faster it will move. As result, the oligonucleotides will be arranged from smallest to largest, reading the gel from bottom to top.
Gel Analysis and Determination of DNA Sequence
The last step simply involves reading the gel to determine the sequence of the input DNA. Because DNA polymerase only synthesizes DNA in the 5' to 3' direction starting at a provided primer, each terminal ddNTP will correspond to a specific nucleotide in the original sequence (e.g., the shortest fragment must terminate at the first nucleotide from the 5' end, the second-shortest fragment must terminate at the second nucleotide from the 5' end, etc.) Therefore, by reading the gel bands from smallest to largest, we can determine the 5' to 3' sequence of the original DNA strand.
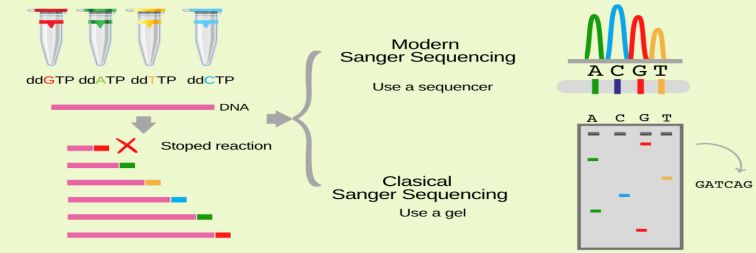 Figure 3. Overview Sanger sequencing
Figure 3. Overview Sanger sequencing
Advantages of Sanger Sequencing
Sanger sequencing is a time-tested methodology famed for its high accuracy. This classic technique meticulously pinpoints the order of bases in DNA, particularly adept when applied to relatively short DNA fragments. Renowned for its robustness, the straightforward mechanism and consistent process of Sanger sequencing lead to a high degree of reliability. Given proper operation, it provides steadfast and reproducible outcomes.
Its applicability is broad and adaptable, facilitating projects of various scales, ranging from sequencing singular genes to entire genomes. Hence, it finds ample utilization across research, clinical diagnosis, and other domains. Compared to certain NGS technologies, the associated equipment and reagent costs of Sanger sequencing are notably lower. Consequently, it is an appealing option for those projects dealing with limited budgets. When it comes to sequencing lengthy DNA fragments, Sanger sequencing exhibits superior performance relative to some next-gen counterparts. This is critically important for projects necessitating the sequencing of complete genes or specific regions.
Applications of Sanger Sequencing
Sanger DNA sequencing is widely used for research purposes like (1) targeting smaller genomic regions in a larger number of samples, (2) sequencing of variable regions, (3) validating results from NGS studies, (4) verifying plasmid sequences, inserts, mutations, (5) HLA typing, (6) genotyping of microsatellite markers, and (6) identifying single disease-causing genetic variants.
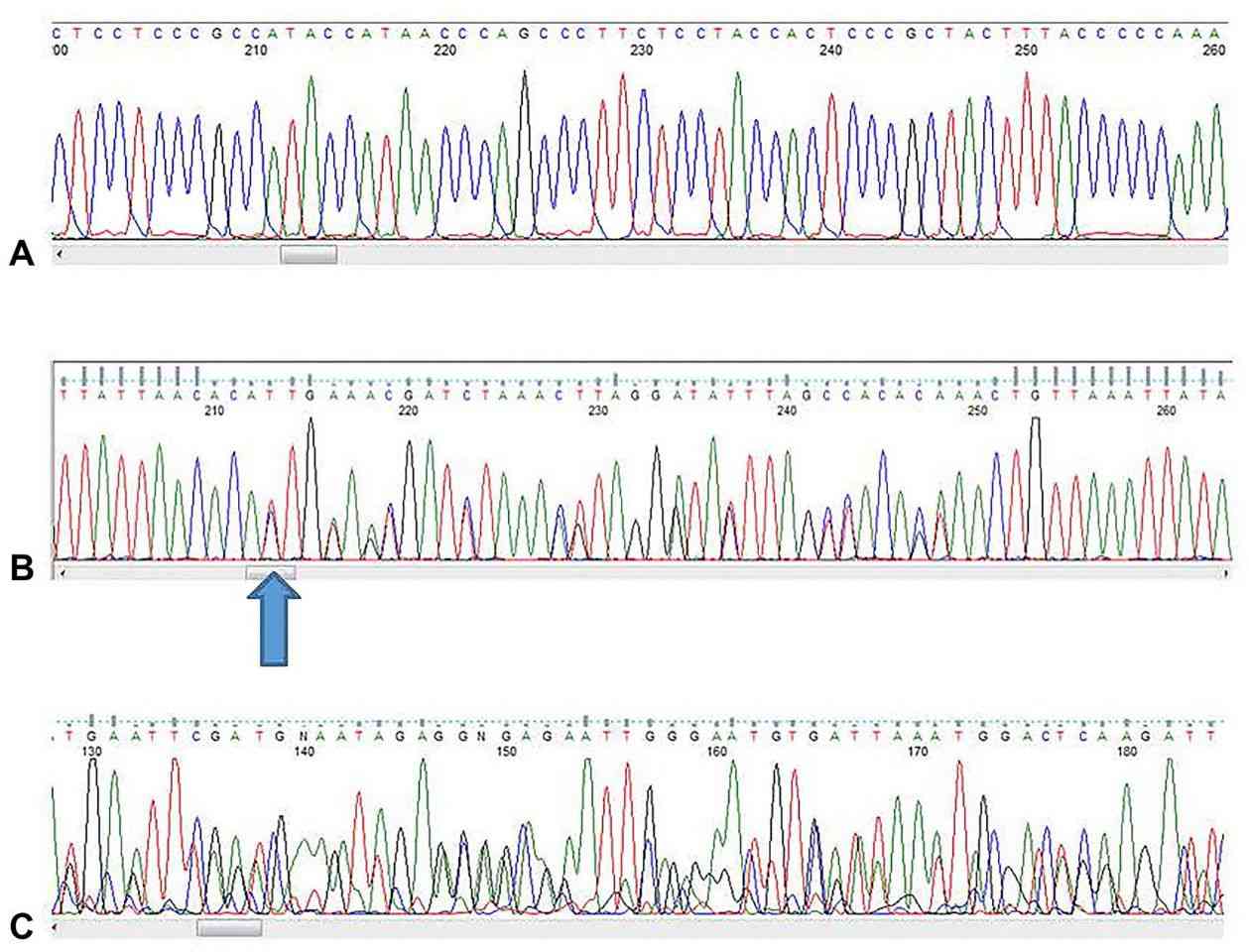 Figure 4. Examples of sequences. A good nucleic acid sequence. (Crossley et al., 2020)
Figure 4. Examples of sequences. A good nucleic acid sequence. (Crossley et al., 2020)
Sanger sequencing is pivotal in many areas of research even today.
In genomics research, this process is vital for gene localization and cloning by identifying, locating genes, and verifying the accuracy of cloned segments. Even though genomics sequencing cannot rival the speed of NGS technologies, Sanger sequencing holds its own when sequencing small genomes or specific gene regions. It also proves helpful in detecting mutations such as Single Nucleotide Polymorphism (SNP) as well as insertions or deletions. In molecular evolution, Sanger sequencing is used for species identification by comparing the gene sequences of known species in order to determine the phylogeny of unknown ones. Through sequencing the DNA of various species, we can unveil the kinship and evolutionary history of many species. Furthermore, it proves instrumental in medical diagnosis for identifying genetic variations related to genetic diseases and detecting oncogene mutations in tumor cells to guide treatment.
In the agricultural and environmental protection sectors, it's used for breed identification by determining the varieties or kinship of crops and livestock. Also, environmental DNA analysis completed through sequencing DNA in environmental samples gives insight into biodiversity and the health of ecological systems. Forensic applications of Sanger sequencing are seen in personal identification by sequencing individual's DNA for judicial identification and personal recognition. DNA fingerprint identification, which makes use of this method, aids in the analysis of DNA evidence in criminal cases and the confirmation of a suspect's identity. Moreover, emergent applications include single-cell sequencing, in conjunction with single-cell technologies for exploring the gene expression and mutation of single cells. The field of epigenetics features Sanger sequencing in identifying epigenetic markers like DNA methylation or histone modification to gain greater insights into epigenetic regulation. In synthetic biology, it is used to verify the accuracy and function of synthetic DNA sequences.
Limitations of Sanger Sequencing
Limited Scope of Application: The Sanger sequencing method is not ideally suited for the analysis of longer DNA sequences or for large-scale sequencing projects. Due to its complexity and cost, it is more appropriate for small-scale projects with specified objectives.
Restricted Sequencing Length: The length of sequences that Sanger sequencing can decode is restricted by the specific dideoxynucleotide triphosphates (ddNTPs) used during the reaction process. This necessitates multiple rounds of reactions and analyses for long fragment sequencing, which in turn, increases the workload and time cost.
Low Throughput: Compared to certain NGS technologies, Sanger sequencing has a relatively lower throughput. Consequently, fewer sequences can be completed within the same time frame, marking it unsuitable for extensive sequencing projects.
Manual Operation Influences Accuracy: Gel electrophoresis analysis and sequence chromatogram interpretation, integral components of the Sanger sequencing process, often require manual operation. This introduces potential subjective judgement and operation errors, consequently affecting the accuracy of the sequencing outcome.
Sanger Sequencing and NGS Comparison
In the discourse concerning Sanger sequencing and next-generation sequencing, we implicitly address the evolution and advancement in DNA sequencing techniques. Sanger sequencing, an early sequencing technique, has been acclaimed for its reliability and precision. Nevertheless, with the course of scientific progression, NGS technologies have emerged, significantly driving the development in the field of DNA sequencing. There are conspicuous differences between Sanger sequencing and NGS technologies in aspects such as their principles, throughput, velocity, costs, range of applications and data analysis.
Principles and Techniques:
Sanger method adopts the chain termination method. This technique employs di-deoxy chain terminators tagged with distinct fluorescence colors, enabling the incorporation of a specific base at each position on the DNA strand.
NGS technologies represent a set of emerging high-throughput sequencing methods, enabling large-scale massively parallel DNA sequencing. These techniques, including Illumina, Ion Torrent, PacBio, and others, allow simultaneous sequencing of numerous DNA fragments, significantly enhancing sequencing speed and efficiency.
Throughput and Speed:
Sanger sequencing is generally limited to handling a modest number of DNA fragments, with a relative slower pace. In contrast, NGS techniques achieve a higher throughput and speed, capable of concurrently processing hundreds of millions to billions of DNA fragments and accomplishing complete genomic sequencing in a significantly shortened duration.
Cost:
Sanger sequencing tends to be relatively expensive, especially with regard to large-scale sequencing projects. NGS technologies, meanwhile, prove to be more cost-efficient due to their capacity to sequence a bulk of DNA fragments at once, therefore, reducing the cost per fragment.
Scope of Application:
Sanger sequencing is typically engaged in smaller-scale sequencing projects, such as sequencing single genes or specific DNA regions. NGS technologies find their extensive applications across various domains including genomics, transcriptomics, and epigenomics, enabling large-scale, in-depth, and high-resolution DNA sequencing.
Data Analysis:
The modest data volume makes the analysis relatively simplistic for Sanger sequencing. However, the substantially larger data volume generated by NGS ordinarily calls for intricate bioinformatics tools and algorithms for processing and interpretation.
Table 1 Comparison of the advantages and disadvantages of Sanger sequencing and NGS
| Sanger Sequencing | Next-Generation Sequencing (NGS) | |
| Advantages |
|
|
| Disadvantages |
|
|
Each of these sequencing approaches, including Sanger sequencing, next-generation sequencing, and third-generation sequencing, has its unique pros and cons. It is advisable to select the method that best suits the research objective and sample under study.
References:
- Crossley BM, Bai J, Glaser A, et al. Guidelines for Sanger sequencing and molecular assay monitoring. Journal of Veterinary Diagnostic Investigation. 2020 Nov;32(6).
- Mardis ER. DNA sequencing technologies: 2006–2016. Nature protocols. 2017 Feb;12(2).
- Saini M K, Gaurav H B, Kumar J, et al. DNA Sequencing techniques: Sanger to Next Generation Sequencing. DNA, 2023, 3(09): 2378-2393.
- Crossley B M, Bai J, Glaser A, et al. Guidelines for Sanger sequencing and molecular assay monitoring. Journal of Veterinary Diagnostic Investigation, 2020, 32(6): 767-775.


 Sample Submission Guidelines
Sample Submission Guidelines
