In the regulatory network of the human immune system, the human leukocyte antigen (HLA) gene family, which encodes the major histocompatibility complex (MHC), plays a central role in regulating immune responses and the genetic susceptibility mechanisms of diseases. HLA molecules mediate adaptive immune response through antigen presentation mechanism, and allelic variation caused by its high polymorphism constitutes the genetic basis of individual immune response differences. Epidemiological and molecular genetic studies show that HLA gene polymorphism is closely related to the occurrence and development of many diseases.
This paper expounds the structure and function of HLA gene family, and the relationship between its polymorphism and the susceptibility to autoimmune diseases, infectious diseases and cancer immunotherapy.
Overview of HLA Gene Family
HLA system is an important part of the human immune system, which is located on the short arm of chromosome 6 and constitutes the MHC. The HLA gene family is famous for its high polymorphism, which is the genetic basis of individual immune response differences and is closely related to the susceptibility to many diseases.
The structure of the HLA gene family is complex and can be divided into class I, class II, and class III regions. Among them, HLA class I and class II genes are directly related to immune response.
- HLA class I genes include HLA-A, HLA-B, and HLA-C sites, and the encoded molecules are widely expressed on the surface of almost all nucleated cells.
- HLA class II genes mainly include HLA-DR, HLA-DQ, and HLA-DP sites, and the encoded molecules are mainly expressed on the surface of antigen-presenting cells (such as macrophages, dendritic cells, and B cells).
The high polymorphism of the HLA gene is one of its most remarkable characteristics. Up to now, there are thousands of alleles at HLA-A, HLA-B, and HLA-DRB1. This polymorphism mainly comes from single nucleotide polymorphism (SNP) in gene sequence, insertion/deletion mutation, and gene recombination. There are significant differences in the frequency distribution of HLA alleles among different races and populations, which provides important clues for studying the relationship between HLA and disease susceptibility.

Structure of HLA Molecules (Chaplin et al., 2010)
Structure and Function of HLA Class I Molecules
HLA class I molecules are heterodimers composed of heavy chain (α chain) and light chain (β2-microglobulin). The α chain is encoded by HLA class I genes and contains three domains (α1, α2, and α3), in which α1 and α2 domains together form a peptide binding groove, which is responsible for binding endogenous antigen peptides. β2-microglobulin is non-polymorphic and encoded by the gene on chromosome 15. Its main function is to stabilize the structure of HLA class I molecules.
The peptide binding groove of HLA class I molecules is highly specific, and different HLA class I alleles can bind antigen peptides with different sequence characteristics. This specificity is determined by the amino acid residues at the bottom and side wall of the peptide binding tank, which form specific "anchor sites" and interact with specific amino acid residues of the antigen peptide. It is precisely because of the high polymorphism of the HLA class I gene that HLA class I molecules of different individuals can bind and present different antigenic peptides, which leads to the difference in immune response among individuals.
HLA Class II Molecules Structure and Function
HLA class II molecules are heterodimers composed of α chain and β chain, both of which are encoded by HLA class II genes, and each contains two domains (α1, α2, and β1, β2). Among them, α1 and β1 domains together form a peptide binding groove, which is responsible for binding exogenous antigen peptides; The α2 and β2 domains are mainly involved in the interaction with T cell surface molecules.
Compared with HLA class I molecules, the peptide binding groove of HLA class II molecules is more flexible and can bind longer antigen peptides (usually 13-17 amino acids). The polymorphism of HLA class II molecules mainly concentrates in α1 and β1 domains, especially in the key region of the peptide binding groove, which enables different HLA class II alleles to bind different antigenic peptides.
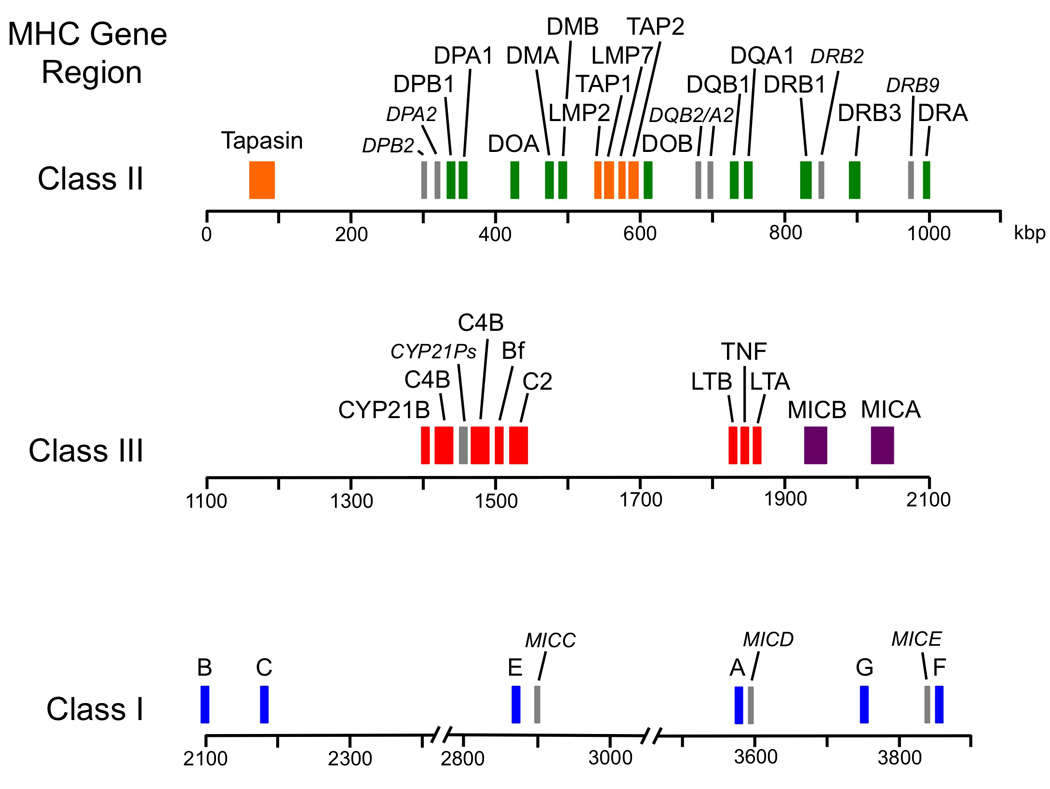
Molecular map of the human major histocompatibility complex (Chaplin et al., 2010)
Interaction Between HLA Molecules and T cell Receptors
The main function of HLA molecules is to present antigen peptides to T cells, thus initiating specific immune responses. T Cell Receptor (TCR) on the surface of T cells can recognize antigen-peptide complexes presented by HLA molecules. This recognition is highly specific and depends not only on the sequence of antigen peptide but also on the type of HLA molecule.
The interaction between HLA molecules and TCR is the basis of the immune system’s specific recognition of antigens. When pathogens invade, antigen-presenting cells will present antigen peptides of pathogens on the cell surface through HLA molecules, and T cells will recognize these antigen peptides-HLA complexes through TCR, thus being activated and initiating immune response. In this process, HLA molecules play a bridge role, transmitting the information of antigen peptides to T cells, thus realizing the specific recognition and elimination of pathogens by the immune system.
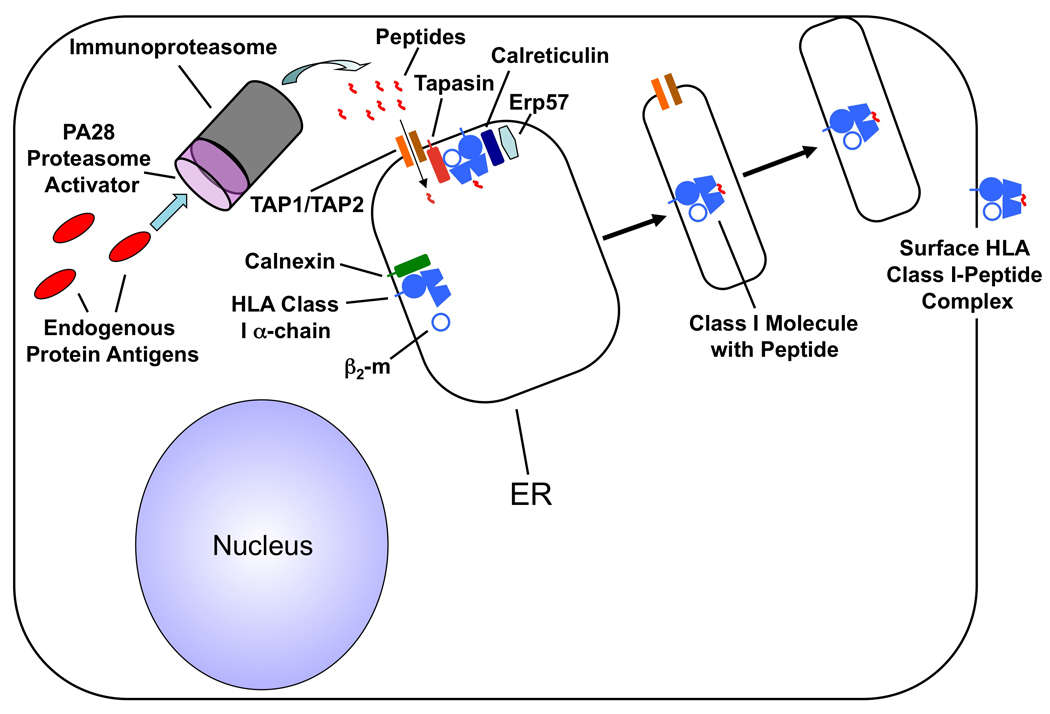
Cellular pathway for processing and presentation of endogenous antigens (Chaplin et al., 2010)
HLA typing technologies have evolved from serological methods to sophisticated molecular approaches, revolutionizing precision medicine and immunology. Early PCR-based techniques like sequence-specific primer (PCR-SSP) and oligonucleotide probe (PCR-SSO) enabled medium-resolution genotyping, while Sanger sequencing set the gold standard for accuracy. Today, next-generation sequencing (NGS) dominates, offering high-throughput, full-gene coverage and single-nucleotide resolution, critical for transplant matching and rare allele discovery.
Association Between HLA Typing and Autoimmune Diseases
Autoimmune diseases refer to diseases caused by the body’s immune system mistakenly attacking its tissues and organs. A large number of studies have shown that HLA genes are closely related to the susceptibility to various autoimmune diseases. The molecular mechanism of this association may be related to the ability of HLA molecules to present some autoantigen peptides. When HLA molecules can present some autoantigen peptides efficiently, it may lead to the activation of autoreactive T cells, which may lead to autoimmune diseases.
HLA-B27 and Ankylosing Spondylitis
Ankylosing Spondylitis (AS) is a chronic inflammatory disease that mainly affects the spine and sacroiliac joints, and its incidence rate in the population is about 0.1-1%. A large number of studies show that HLA-B27 has a strong correlation with the susceptibility of AS. In different races and populations, the risk of AS of HLA-B27 positive individuals is 20-100 times higher than that of HLA-B27 negative individuals.
The molecular mechanism of the association between HLA-B27 and AS has not been fully clarified.
- One hypothesis is that HLA-B27 may wrongly present some bacterial antigen peptides, which have similar structures to human self-antigens, thus triggering cross-reactions and leading to autoimmune attacks.
- Another hypothesis holds that the structure of the HLA-B27 molecule itself may make it more prone to misfolding, which may lead to endoplasmic reticulum stress and inflammatory reaction.
Recent studies have also found that there may be differences in susceptibility and clinical manifestations between different subtypes of HLA-B27 and AS. For example, HLA-B27:05 subtype is closely related to AS in most people, while other subtypes, such as HLA-B27:09, have weak or no association with diseases in some people. This difference in subtype level provides a new perspective for further understanding the relationship between HLA-B27 and AS.
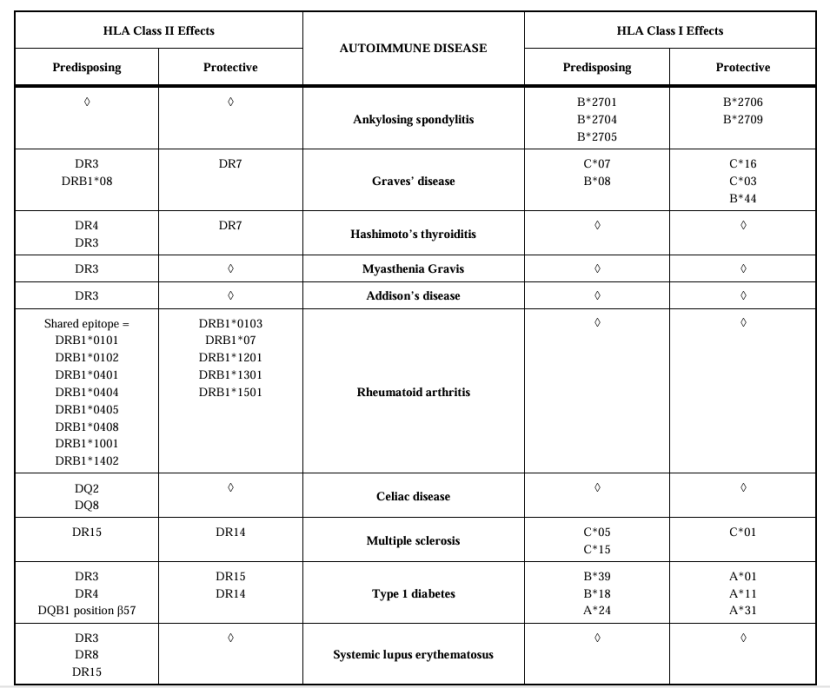
Summary of the Major Associations Within the HLA Class II and Class I Region with Common Autoimmune Diseases (Gough et al., 2007)
HLA-DR4 and Rheumatoid Arthritis
Rheumatoid Arthritis (RA) is a common autoimmune disease, which mainly affects joints, leading to chronic inflammation, joint destruction, and dysfunction. Studies have shown that HLA-DR4 (especially the HLA-DRB1*04 allele) is closely related to the susceptibility to RA. In the Caucasian population, about 70-80% of patients with RA carry the HLA-DR4 allele, while the carrying rate in the normal population is about 30-40%.
The molecular mechanism of the association between HLA-DR4 and RA may be related to the ability of HLA-DR4 molecules to present their antigenic peptides. It is found that the peptide binding groove of the HLA-DR4 molecule has specific structural characteristics, which can efficiently bind and present some autoantigen peptides with specific amino acid sequences, thus activating autoreactive T cells and triggering immune attacks.
Further research found that some amino acid residues in the peptide binding groove of HLA-DR4 molecules are closely related to the susceptibility to RA. The region where these residues are located is called "Shared Epitope" and exists in alleles such as HLA-DRB104 and HLA-DRB101. Individuals with shared epitopes have a significantly increased risk of RA, and the number of shared epitopes may be related to the severity of the disease.
HLA Protection/Risk Factors in Infectious Diseases
The HLA gene also plays an important role in infectious diseases. On the one hand, some HLA alleles may give individuals resistance to specific pathogens and become protective factors. On the other hand, some HLA alleles may increase the susceptibility of individuals to specific pathogens and become risk factors. This association between HLA and susceptibility to infectious diseases is mainly due to the difference in the ability of HLA molecules to present pathogen antigen peptides.
Association Between HIV Infection and HLA Types
Human Immunodeficiency Virus (HIV) infection can lead to acquired immunodeficiency syndrome (AIDS). A large number of studies show that the HLA gene is closely related to the progress and prognosis of HIV infection.
Some HLA alleles are related to the slow progress of HIV-infected diseases and become protective factors. For example, HLA-B57 and HLA-B27 alleles have been proven to be associated with slower disease progression after HIV infection in many studies. Individuals carrying these alleles often take longer to develop AIDS after being infected with HIV, and the mechanism may be related to the fact that these HLA molecules can efficiently present HIV antigen peptides, thus inducing a stronger antiviral immune response.
On the contrary, some HLA alleles are related to the rapid disease progression after HIV infection and become risk factors. For example, HLA-B35 and HLA-A25 alleles were found to be associated with rapid disease progression after HIV infection. These HLA molecules may not be able to effectively present the antigenic peptides of HIV, resulting in insufficient antiviral immune response, rapid replication of the virus in the body, and accelerated disease progress.
The HLA gene not only affects the disease progression of HIV infection but also is related to the effect of antiretroviral therapy. It is found that some HLA alleles may be related to the adverse reactions of antiretroviral drugs, such as HLA-B*57:01, which is closely related to the hypersensitivity caused by Abacavir, which is also one of the important applications of HLA typing in clinical medication guidance.
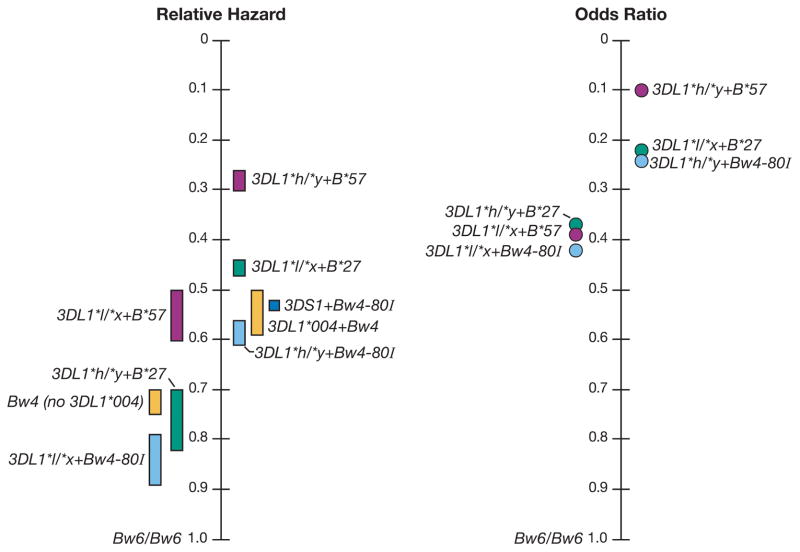
KIR3DL1 + HLA-B Bw4 continuum of HIV viral load control (Martin et al., 2013)
Association Between Hepatitis B Infection and HLA Types
Hepatitis B Virus (HBV) infection is a global health problem, which can lead to acute and chronic hepatitis, liver cirrhosis, and even liver cancer. Studies have shown that the HLA gene is closely related to the outcome and prognosis of HBV infection.
In the natural history of HBV infection, the individual’s immune response-ability is the key factor to determine the outcome of infection. As the key molecule of antigen presentation, HLA polymorphism may affect the recognition and response of the body to HBV antigen. It is found that some HLA alleles are related to the spontaneous clearance ability after HBV infection, such as HLA-A11 and HLA-B13 alleles, which have been found to be related to HBV clearance in some studies and become protective factors.
On the contrary, some HLA alleles are related to the susceptibility to chronic HBV infection. For example, the HLA-DR3 allele was found to be associated with an increased risk of chronic HBV infection. Individuals who carry these alleles may have an insufficient immune response to HBV antigen, which may lead to the virus being unable to be effectively eliminated, thus developing into chronic infection.
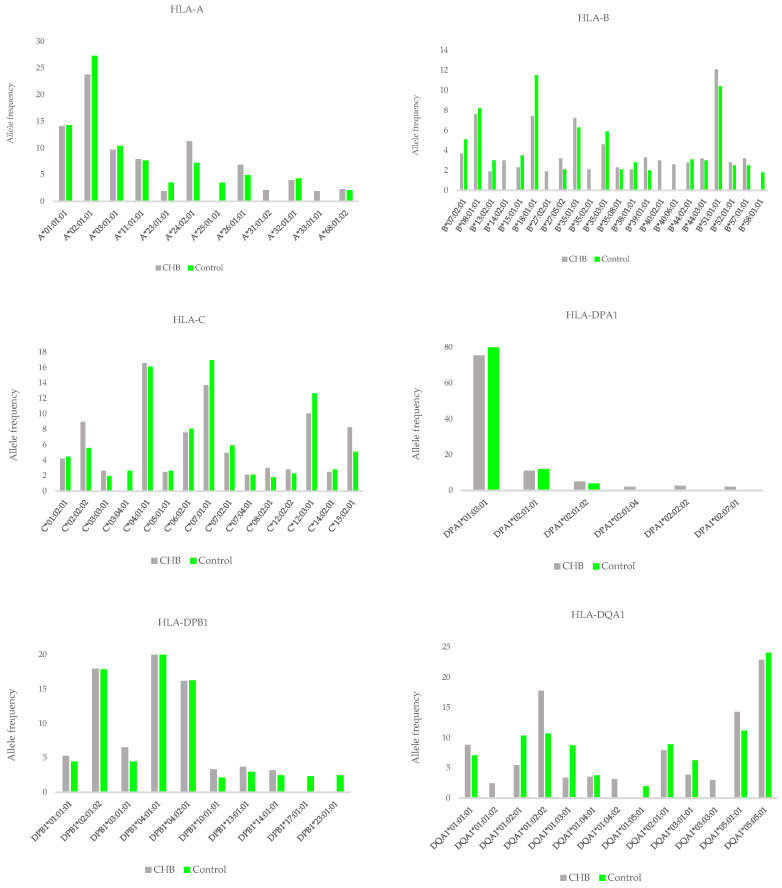
Frequency distribution of HLA alleles in the CHB group and the control group (Tălăngescu et al., 2024)
Relationship Between HLA and Cancer Immunotherapy
Cancer immunotherapy has been a breakthrough in the field of tumor treatment in recent years, among which checkpoint inhibitor therapy is particularly eye-catching. More and more studies show that the HLA gene is closely related to the efficacy of checkpoint inhibitors, which provides a new direction for personalized cancer immunotherapy.
HLA Dependence of the Efficacy of Checkpoint Inhibitors
Checkpoint inhibitors reactivate the anti-tumor immune response of T cells by releasing the inhibition of tumor cells in the immune system. However, not all patients can benefit from the treatment of checkpoint inhibitors, and there are significant individual differences in their efficacy. It is found that this individual difference may be closely related to the HLA type of patients.
HLA molecules play a key role in anti-tumor immune response. HLA molecules on the surface of tumor cells can present tumor antigen peptides for T cells to recognize. Therefore, the expression and functional status of HLA molecules directly affect the ability of T cells to recognize and attack tumor cells. Studies have shown that the loss or abnormality of HLA class I molecule expression is one of the important mechanisms for tumor cells to escape immune surveillance.
The mechanism of HLA dependence may involve many aspects. On the one hand, different HLA alleles may affect the presentation efficiency and types of tumor antigen peptides, thus affecting the activation of T cells and the intensity of the anti-tumor immune response. On the other hand, HLA molecules may interact with the mechanism of checkpoint inhibitors.
HLA and Tumor Mutation Load
Tumor Mutation Burden (TMB) is one of the important biomarkers to predict the efficacy of checkpoint inhibitors. Generally speaking, tumor patients with higher TMB have a higher response rate to checkpoint inhibitors. It is found that there may be a correlation between the HLA gene and TMB.
Polymorphism of HLA molecules may affect the body’s recognition and response to tumor mutant antigens. On the one hand, some HLA alleles may be more conducive to presenting tumor mutant antigens, thus making tumors with high TMB more easily recognized and attacked by the immune system. On the other hand, the expression of HLA molecules may affect the sensitivity of tumor cells to immune attack, thus indirectly affecting the relationship between TMB and curative effect.
In addition, the mutation of the HLA gene itself may also affect the immunogenicity of the tumor. It is found that the mutation of the HLA gene may lead to abnormal expression or functional defects of HLA molecules, thus affecting the presentation of tumor antigens, reducing the immunogenicity of tumors, and leading to poor therapeutic effect of checkpoint inhibitors.
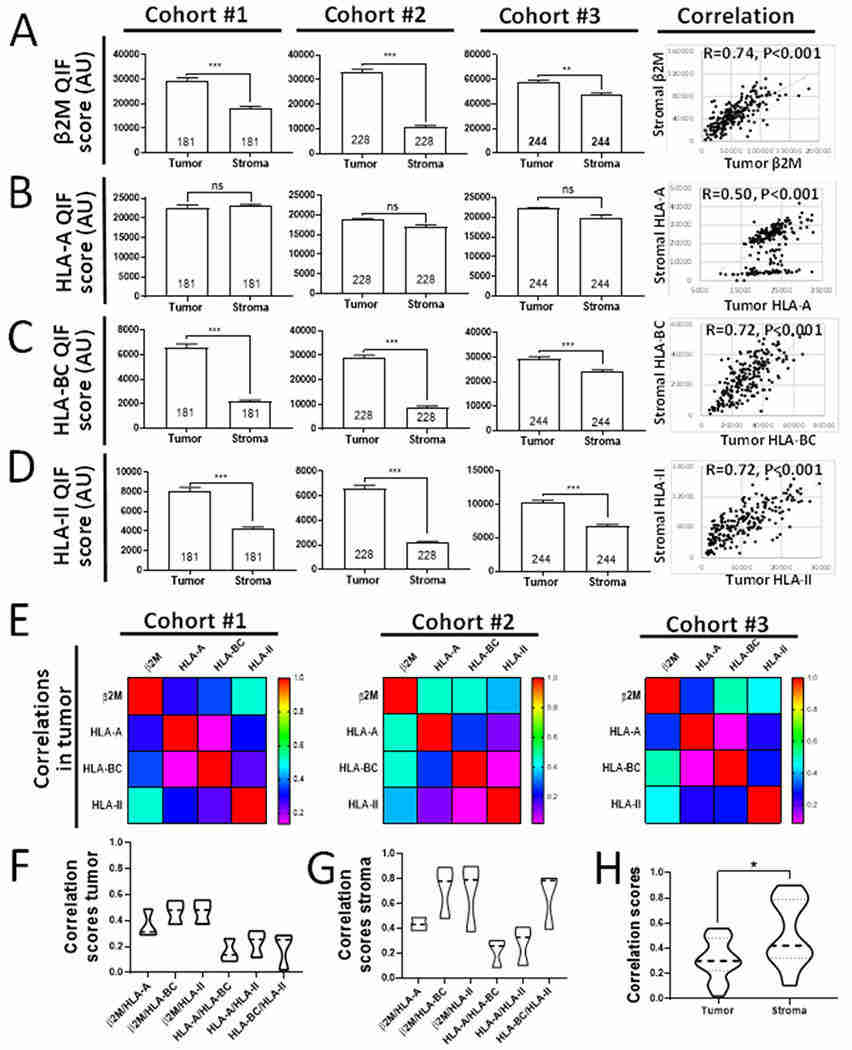
Cancer and stromal cell specific distribution and associations between HLA class-I and HLA class-II subunits in NSCLC (Datar et al., 2021)
Conclusion
To sum up, the HLA gene is a key component of the human immune system, and its polymorphism is closely related to the susceptibility to many diseases. HLA types play an important role in autoimmune diseases, infectious diseases, and cancer immunotherapy. In-depth study on the relationship between HLA and diseases not only helps to clarify the pathogenesis of diseases but also provides new ideas and targets for the prevention, diagnosis, and treatment of diseases.
Learn More
References:
- Chaplin DD. "Overview of the immune response." J Allergy Clin Immunol. 2010 125(2 Suppl 2): S3-S23 https://doi.org/10.1016/j.jaci.2009.12.980
- Gough SC, Simmonds MJ. "The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action." Curr Genomics. 2007 8(7): 453-465 https://doi.org/10.2174/138920207783591690
- Tălăngescu A, Tizu M., et al. "HLA Genetic Diversity and Chronic Hepatitis B Virus Infection: Effect of Heterozygosity Advantage." Med Sci (Basel). 2024 12(3): 44 https://doi.org/10.3390/medsci12030044
- Martin MP, Carrington M. "Immunogenetics of HIV disease." Immunol Rev. 2013 254(1): 245-264 https://doi.org/10.1111/imr.12071
- Datar IJ, Hauc SC, Desai S., et al. "Spatial Analysis and Clinical Significance of HLA Class-I and Class-II Subunit Expression in Non-Small Cell Lung Cancer." Clin Cancer Res. 2021 27(10): 2837-2847 https://doi.org/10.1158/1078-0432.ccr-20-3655


 Sample Submission Guidelines
Sample Submission Guidelines