Prokaryotes such as bacteria and archaea tend to exhibit simplicity in their cellular structure, devoid of membrane-enclosed organelles like ribosomes, endoplasmic reticulum, and mitochondria. In contrast, eukaryotes, encompassing both single cell organisms and their multicellular derivatives, boast significantly more complex cellular architectures. In terms of genetic material organization, prokaryotes typically feature more rudimentary ribosomes, whereas eukaryotes possess nuclei encapsulated by nuclear membranes.
What are Ribosomes?
Ribosomes are indispensable components of biological processes, responsible for protein synthesis within an organism. These ribosomes, distributed in the cytoplasm and rough endoplasmic reticulum, comprise two subunits - a large one and a small one. The structure and function of ribosomes exhibit variations when comparing prokaryotes and eucharyotes.
Prokaryotic ribosomes consist of two subunits, namely 50S and 30S, which combine to form a 70S ribosome. Bacterial ribosomal RNA (rRNA) is categorized into three types based on sedimentation coefficients: 5S, 16S, and 23S rRNA. Among these, the 16S rRNA plays a pivotal role within the smaller subunit of prokaryotic species. The sequence information afforded by the 16S rRNA is instrumental in examining phylogenetics and classifications of organisms, hence it is ubiquitously employed in studies pertaining to microbial ecology and phylogenetics.
Eukaryotic ribosomes, on the other hand, comprise 60S and 40S subunits that amalgamate to form an 80S ribosome. Fungal rRNA is characterized into four types according to the sedimentation coefficients: 5S, 5.8S, 18S, and 28S rRNA.
What are 16S, 18S and ITS
The sequence length of 16S rRNA is approximately 1542 bp, encompassing 10 conserved regions and 9 variable regions (V1 to V9). These conserved regions reflect phylogenetic relationships between species, while the variable regions highlight inter-species differences. Particularly noteworthy are the V3, V4, and V5 regions, characterized by high specificity and comprehensive database information, rendering them the optimal choices for bacterial diversity analysis and annotation.
The process of eukaryotic ribosome formation involves the employment of 18S rRNA, a component that forms the small subunit of these ribosomes. Remarkably, this shares a significant degree of homology with the 16S rRNA utilized by prokaryotes. Intricately, this sequence is formulated with a combination of conserved and variable regions, numbering from V1 to V9.
Moreover, eukaryotes harbor a structural entity known as the Internal Transcribed Spacer (ITS) within their rDNA genes. The ITS segregates into two distinct regions: ITS1 and ITS2. Notably, the ITS1 sequence resides between the 18S rRNA and 5.8S rRNA genes, whereas the ITS2 sequence is sandwiched between the 5.8S rDNA and the 28S rDNA genes.
Microbial Amplicon Sequencing and Its Advancements
As an advanced technique, amplicon sequencing, also known as microbial diversity sequencing, leverages next-generation or long-read sequencing platforms to sequence the polymerase chain reaction (PCR) products of specific regions such as 16S rRNA, 18S rRNA, ITS, and functional genes. This approach transcends the limitations of traditional microbiological culture methods, enabling direct extraction of information from environmental samples regarding microbial community structure, evolution, and correlations between microbes and their environment.
The progression from next-generation to long-read sequencing technologies has seen the widespread application of amplicon sequencing across numerous research spheres, particularly in the study of microbial diversity and community structure. Second-generation sequencing platforms, such as the Illumina method, offer the prowess of high-speed, large-scale production of short read DNA fragments, thus enabling the extensive sequencing of amplified subunits. While second-generation sequencing technologies do kapitulate high throughput and precision, their comparative shorter read-length may pose certain constraints for the study of longer amplicon sequences.
long-read sequencing technologies, exemplified by platforms like PacBio, boast notable advantages in long-read capabilities, affording us the ability to acquire extended amplicon sequences. Furthermore, the single-molecule sequencing approach inherent in third-generation sequencing directly retrieves sequencing information from DNA templates, providing heightened flexibility across various research applications. This unique feature enhances the adaptability of third-generation sequencing technologies in diverse scientific contexts.
| Product | Research Targets |
|---|---|
| 16S sequencing | Bacteria/Archaea |
| 18S sequencing | Eukaryotic microorganisms |
| ITS sequencing | Fungi |
| Functional gene amplicon sequencing | Other target genes |
Microbial Full-Length Amplicon Sequencing VS Next-generation Amplicon Sequencing
The PacBio platform, renowned for its extended read lengths, exhibits no GC bias and incorporates automatic error correction through Circular Consensus Sequencing (CCS), ensuring heightened data accuracy.
Demonstrating superior sensitivity, this technology excels in providing more abundant and precise species information by aligning to individual highly variable regions.
Elevating the precision of microbial composition identification, it faithfully reconstructs the microbial community structure within samples.
Accurately discerning the full-length copy polymorphism of 16S, it identifies strain-level variations, thereby enhancing taxonomic resolution.
Achieving accurate sequencing of all highly variable regions from V1 to V9, this methodology truly refines resolution to the species level, consequently bolstering the credibility of annotations.
| Product | Sequencing Platform | Sequencing Volume | PCR Amplification | Barcode Sequences | Research Level | Average Read Length | Accuracy |
| Next-generation | Illumina NovaSeq PE250 | 5w tags start | Required | Required | Genus | 2 * 250bp | Q30 |
| Long-read Amplicon | PacBio Sequel IIe Revio | 1w CCS start | Typically not required | Required | Species | 10-20kb | Q50 |
Microbial Amplicon Sequencing VS Other Microbial Detection Methods
While amplicon sequencing technology finds extensive application in the study of microbial communities, it is essential to recognize that other microbial detection methods exhibit notable advantages in specific aspects. In the subsequent discussion, we will juxtapose amplicon sequencing technology with other prevalent microbial detection methods.
Cultivation Methods: Traditional cultivation methods typically rely on specific nutrient media and environmental conditions to cultivate microorganisms. In comparison to amplicon sequencing, the advantage of traditional cultivation lies in the direct observation and analysis of the morphology, physiology, and biochemical characteristics of live microorganisms. However, the limitation of traditional cultivation methods is evident in their ability to detect only a fraction of cultivable microorganisms, failing to represent the full diversity of the microbial community.
Environmental DNA (eDNA) Detection: Environmental DNA refers to the genetic material of microorganisms or other organisms extracted from environmental samples. This approach enables the direct detection of microbial presence in the environment and is particularly applicable to the study of rare or difficult-to-culture microorganisms. However, eDNA detection may not furnish as comprehensive microbial community structure information as amplicon sequencing and is susceptible to influences from environmental and technical factors.
Microbial Whole Genome Sequencing (WGS): Whole Genome Sequencing is a methodology that involves sequencing all DNA present in environmental samples to acquire comprehensive information about biological functions and genes. This approach allows for a more in-depth analysis of microbial communities at the functional and genetic levels and contributes to the discovery of new species and genes. However, in comparison to amplicon sequencing, Whole Genome Sequencing generates larger datasets, involves relatively complex data processing and analysis, and incurs higher costs.
In comparison with metagenomic shotgun sequencing, please refer to the article Amplicon-Based Next-Generation Sequencing vs. Metagenomic Shotgun Sequencing 16S rRNA Sequencing VS Shotgun Sequencing for Microbial Research ITS Sequencing in Microbiological Research: Introduction, Bioinformatics, and Applications MicroSEQ Identification by 16S/D2 Region Sequencing: Introduction, Protocol, and Applications
Amplicon Sequencing Workflow and Data Analysis
(1) Technical Workflow
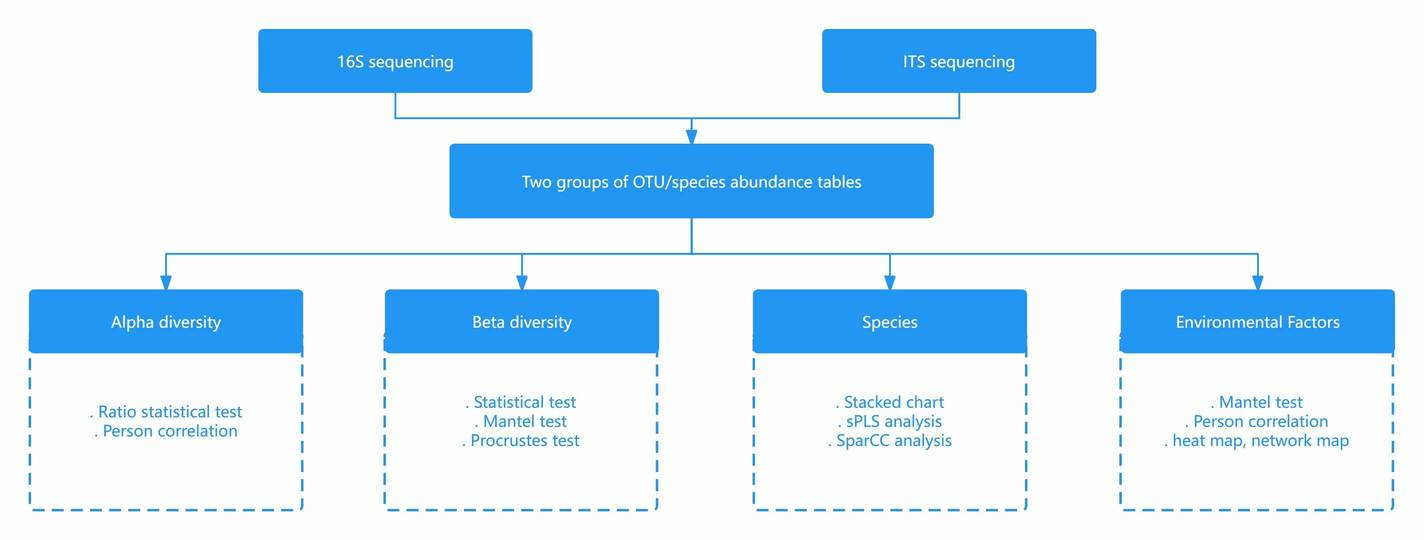
The workflow for amplicon sequencing involves the initial enrichment of specific regions through PCR amplification of total DNA extracted from environmental samples, followed by library construction and sequencing. Subsequently, data undergoes quality control and bioinformatics analysis to unravel the diversity of microbial communities within the environment.
For an in-depth understanding of full-length amplicon sequencing principles, please refer to the introduction on Full-Length Amplicon Sequencing Technology.
![]()
(2) Data Analysis
As amplicon sequencing technology becomes increasingly prevalent in biological research, the processing and analysis of data have emerged as critical stages. To ensure robust scientific outcomes, it is imperative to execute precise quality control measures, eliminate noise and errors from the raw sequencing data, and ultimately derive biological information that aligns with the research objectives.
Case Studies of Amplicon Sequencing Applications
Through the utilization of amplicon sequencing technology, we gain the ability to investigate the diversity, structure, and functionality of microbial communities in diverse environments. The following presents a partial analysis of case studies applying amplicon sequencing in practical scenarios.
Soil Microbial Community Research
Microbial resources in soil play a pivotal role in plant growth and soil fertility. Amplicon sequencing technology facilitates an in-depth exploration of the structure, functionality, and interrelationships within soil microbial communities. This analysis extends to examining their impact on environmental factors, agricultural management practices, and ecosystem functions. The application of this technology contributes robust scientific insights that can underpin sustainable agricultural development and soil ecological restoration.
Abundant fungi dominate the complexity of microbial networks in soil of contaminated site: High-precision community analysis by full-length sequencing
In January 2022, an article addressing the complexity of microbial communities in contaminated soil using full-length amplicon sequencing was published in the "Science of the Total Environment" journal. This study elucidates the composition and dynamic characteristics of microbial communities in polluted soil from a full-length sequencing perspective. Employing both the PacBio and Illumina platforms for simultaneous 16S rRNA/ITS amplicon sequencing, the research scrutinized the structure of microbial communities in contaminated soil. The findings indicate that heavy metals and soil texture significantly influence the microbial community structure. Bacterial communities and dominant taxa are primarily governed by deterministic processes, while other taxa are subject to stochastic processes. Furthermore, the study suggests that with the advancement of databases and improvements in sequencing methodologies, the application of full-length sequencing to environmental samples holds promising prospects.
Aquatic Microbial Community Research
Amplicon sequencing technology holds extensive value in the study of microbial communities in aquatic environments, including seawater, lakes, and rivers. Through in-depth analysis of the composition and dynamic changes in microbial communities within water bodies, we can gain deeper insights into the health of aquatic ecosystems and the current status of environmental conservation. The application of this technology contributes to a better understanding and preservation of aquatic ecosystems, providing a scientific foundation for environmental conservation and sustainable development.
Full-length 16S rRNA gene sequencing reveals spatiotemporal dynamics of bacterial community in a heavily polluted estuary, China
In January 2021, an article was published in the "Environmental Pollution" journal, delving into the spatiotemporal dynamics of bacterial community structure in a heavily polluted estuary revealed through full-length 16S rRNA sequencing. The study employed full-length amplicon sequencing technology to investigate bacterial community structures in the Liao River estuary. Initially, 57 samples were collected and categorized based on environmental conditions, regions, seasons, and lifestyles.
The findings indicated that in marine water, both particle-attached bacteria (PA) and free-living bacteria (FL) exhibited higher α-diversity in the coastal area during the dry season, shifting to the midstream region during the rainy season. Differences in bacterial community structure at the genus level were observed between sediment and seawater samples in coastal areas, with habitat type being the primary influencing factor. Bacterial communities of different lifestyles showed significant differences during the dry season, while no apparent disparities were observed during the rainy season, signifying a consequence of shifts in the lifestyle of planktonic bacteria.
Spatial variations in bacterial community structure were observed in sediment samples, with no seasonal changes. Tides and nutrients emerged as principal drivers for bacterial communities in marine water. This study contributes to understanding the impact of human activities on bacterial communities in the Liao River estuary.
Research on the Human Microbiome
Within the human body, numerous microbial communities exist, such as the gut microbiota and skin microbiota. These microbes play a crucial role in maintaining human health. Through the application of amplicon sequencing technology, we can gain profound insights into the distribution and functions of these microorganisms within the human body. This knowledge serves as vital reference information for the diagnosis, treatment, and prevention of diseases.
High-Resolution Differentiation of Enteric Bacteria in Premature Infant Fecal Microbiomes Using a Novel rRNA Amplicon
identification of intestinal bacteria in the fecal microbiome of preterm infants using amplicon sequencing. The study employed a novel sequencing kit, combining DNA extraction, PCR amplification of the rRNA operon region, and downstream bioinformatics data analysis. Longitudinal microbiome samples from twins admitted to two different Neonatal Intensive Care Units (NICUs) were analyzed using 2500-base pair amplicons spanning the 16S and 23S rRNA genes and aligned to a newly customized 16S-23S rRNA database.
The use of DADA2-inferred Amplicon Sequence Variants (ASVs) provided sufficient resolution to distinguish rRNA variants. These strains were closely related to Klebsiella pneumoniae, Escherichia coli, and Enterobacter strains, not previously sequenced, constituting the first batch of bacteria colonizing the infant intestines post-NICU admission. Different ASV populations were monitored over time between the twins, demonstrating the potential to trace the sources and transmission of symbionts and pathogens. The high-resolution classification obtained through long amplicon sequencing enables the establishment and tracking of microbial communities in infant gut environments within the hospital setting, both temporally and spatially.
Microbial Community Studies in Food and Fermentation Industries
In the realms of food fermentation and the production of bioproducts, microorganisms play a pivotal role in enhancing food quality and safety. The application of amplicon sequencing technology contributes significantly to optimizing fermentation processes, improving product quality, and consequently enhancing the economic efficiency of the industry.
PacBio sequencing reveals bacterial community diversity in cheeses collected from different regions
In January 2021, the Journal of Dairy Science published a study employing PacBio SMRT sequencing technology to investigate the bacterial communities in cheeses collected from various regions. A total of 144 bacterial genera were identified, including Lactobacillus, Streptococcus, Micrococcus, and Staphylococcus, encompassing 217 bacterial species such as lactococci, thermophilic streptococci, Mannheimia haemolytica, and Streptococcus agalactiae. Significant variations among samples were observed at the OTU level, and based on flavor quality detected by an electronic nose system, samples clustered into two distinct groups on a PCA plot. Identification of different bacterial species and metabolic pathways suggested potential factors contributing to the unique flavors of the cheese samples.
Future Prospects and Challenges in Amplicon Sequencing
Amplicon sequencing technology has made significant strides in fields such as environmental science, biology, and medicine. However, numerous challenges persist. To enhance sequencing accuracy and throughput, further advancements and refinement in bioinformatics methodologies are imperative. Additionally, the establishment of more rigorous experimental designs and analytical workflows is necessary to propel continuous growth and progress in this field. Researchers, therefore, must collaboratively address these issues, seeking solutions to propel the continued development of amplicon sequencing technology.
References
- Hongxia M, Jingfeng F, Jiwen L, et al. Full-length 16S rRNA gene sequencing reveals spatiotemporal dynamics of bacterial community in a heavily polluted estuary, China. Environmental Pollution, 2021, 275: 116567.
- Hongxia M, Jingfeng F, Jiwen L, et al. Full-length 16S rRNA gene sequencing reveals spatiotemporal dynamics of bacterial community in a heavily polluted estuary, China. Environmental Pollution, 2021, 275: 116567.
- Graf J, Ledala N, Caimano M J, et al. High-resolution differentiation of enteric bacteria in premature infant fecal microbiomes using a novel rRNA amplicon. MBio, 2021, 12(1): 10.1128/mbio. 03656-20.
- Yang C, Zhao F, Hou Q, et al. PacBio sequencing reveals bacterial community diversity in cheeses collected from different regions. Journal of Dairy Science, 2020, 103(2): 1238-1249.