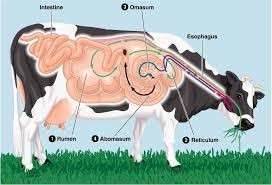
Importance of Rumen Microbiota
The rumen microbial community comprising of archaea, bacteria, fungi, and ciliated protozoa is characterized by high population density, remarkable diversity, and complex microecological interactions. Among ruminal microbes, rumen bacteria accounting for 1011 cells/ml of rumen content are the most diverse group. The rumen microbiota is an indispensable part of ruminants shaping their nutrition and health. However, only 15-20% of the rumen microorganisms can be identified and characterized by conventional culturing and microscopy. The emerging next-generation sequencing (NGS) and long-read sequencing techniques provide a rapid, reproducible, comprehensive tool for the qualitative and quantitative assessment of microbial diversity in rumen ecosystem.
Accelerate Research and Practice in Rumen Microbiota
Based on our NGS, PacBio SMRT sequencing, and Nanopore sequencing platforms, we can help you identify and study the microbes present in the rumen, including their relative abundance, diversity, function, and evolutionary relationships. We also simultaneously study rumen microbiota and metabolic phenotypes to identify the associations between ruminal microbes and the nutrition and health of ruminants, and to discover potential biomarkers of digestive functions. This service is of great importance to a range of fields including scientific research, agricultural practices, and the development of supplements and therapies.
The Main Research Directions
- Composition and similarity of rumen microbiota of ruminants in individual animals.
- Effects of certain treatment on rumen microbiota and rumen fermentation.
- Identify host-microbe associations and potential biomarkers of digestive functions.
What Can We Do?
- 1. Decipher the microbial community structure and diversity in rumen ecosystem
- 2. Quantify ruminal microorganisms
- 3. Explore the rumen microbiota and the metabolic phenotype for identifying host-microbe associations
Note: Our service is for research use only, and not for therapeutic or diagnostic use.
Detectable Objects
Gastrointestinal tract content samples of ruminants, like cattle, bison, and buffalo (bovines), goats, sheep, deer, alpacas, llamas, guanacos.
Detectable Microorganisms
Bacteria, archaea, fungi, protozoa, etc.
Technical Platforms
We can perform 16S/18S/ITS sequencing or metagenomics on Illumina Hiseq/Miseq, PacBio SMRT systems, or Nanopore systems according to your needs. Additionally, we own PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time PCR, Clone library, and other technology platforms.
Sample Requirements
-

- DNA sample: DNA > 500 ng, concentration > 10 ng/ul, and OD260/280 = 1.8-2.0.
- Ensure that the DNA is intact and non-degraded, avoiding repeated freezing and thawing cycles
- Please use enough dry ice or ice packs during shipment.
Workflow
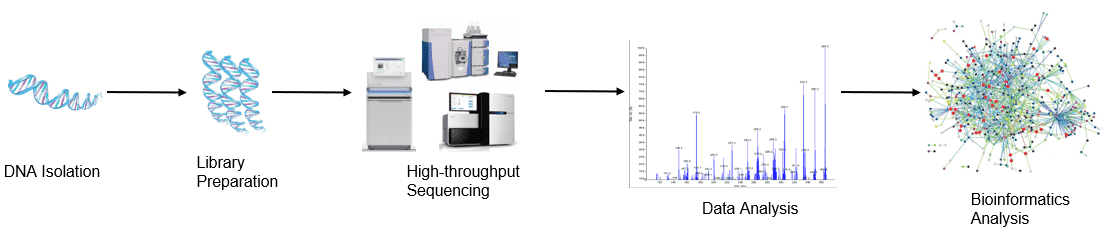 Figure 1. High-throughput sequencing analysis process.
Figure 1. High-throughput sequencing analysis process.
 Figure 2. PCR-DGGE analysis process.
Figure 2. PCR-DGGE analysis process.
Bioinformatics Analysis
| Basic Analysis | Routine Analysis (According to Customer Requirements) | Advanced Data Analysis |
|---|---|---|
| Sequence Filtering and Trimming | Heatmap | Phylogenetic Tree |
| Sequence Length Distribution | VENN | LDA-Effect Size (LEfSe) |
| OTU Clustering and Species Annotation | Principal Components Analysis (PCA) | Network Analysis |
| Diversity Index | Microbial Community Structure Analysis | Correlation Analysis |
| Shannon-Wiener Curve | α Diversity Index Analysis | |
| Rank-Abundance Curve | Matastats Analysis | |
| Dilution Curve | Weighted Unifrac test | |
| Multiple Contrast | CCA/RDA Analysis | |
| Heatmap | ||
| Principal Components Analysis (PCA) |

