Backgrounds
Plasmids are key genetic elements in bacteria, providing traits like antibiotic resistance and virulence factors. Their spread and stability are influenced by growth costs, transfer rates, and evolutionary changes. This study examines whether the variation in plasmid stability traits and their evolution during the absence of antibiotics affect the persistence of resistance plasmids. Using clinical E. coli strains and their associated plasmids, researchers tracked plasmid frequencies and stability traits, revealing that rapid evolution can alter plasmid dynamics, often overriding initial stability traits and demonstrating strain-specific evolutionary patterns.
Materials & Methods
Method:
-
WGS
- Illumina MiSeq
- Oxford Nanopore MinION
Results
Whole-genome sequencing revealed that genetic changes in ESBL plasmids and bacterial chromosomes are specific to the strain-plasmid combination. Evolved clones showed parallel mutations linked to increased plasmid transfer rates and interactions with mobile elements. Chromosomal changes, including deletions of pathogenicity islands, varied with the strain and plasmid, affecting bacterial growth.
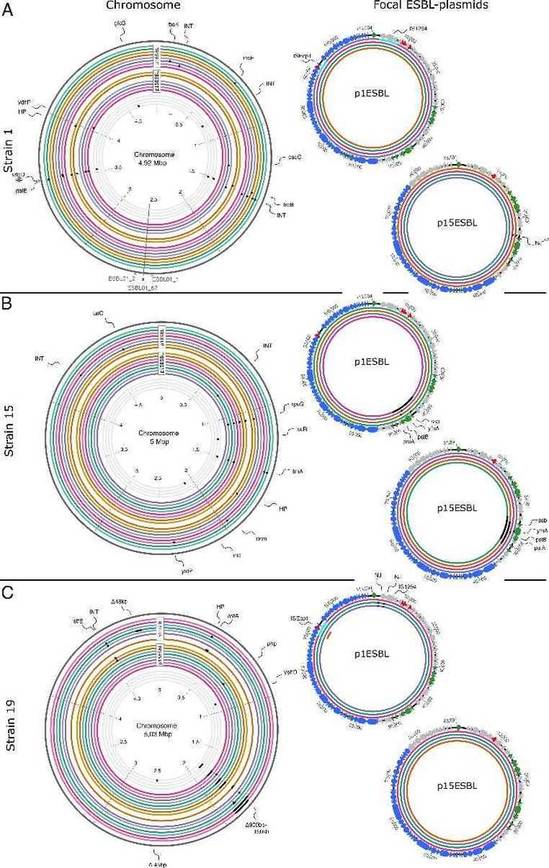 Fig 1. Genetic changes in chromosomes and ESBL plasmids after 15 d of serial passage without antibiotics.
Fig 1. Genetic changes in chromosomes and ESBL plasmids after 15 d of serial passage without antibiotics.
Evolved p1ESBL and p15ESBL plasmids in strain 15 showed increased transfer rates due to mutations in the plasmid leading region, specifically in the ssi3 operon. These mutations, including stop codon SNPs and frame shifts, enhanced transfer rates in strain 15 but not in other strains, indicating a strain-specific effect. The findings suggest that mutations in this region may be a common adaptation mechanism for improving plasmid transfer in clinical settings.
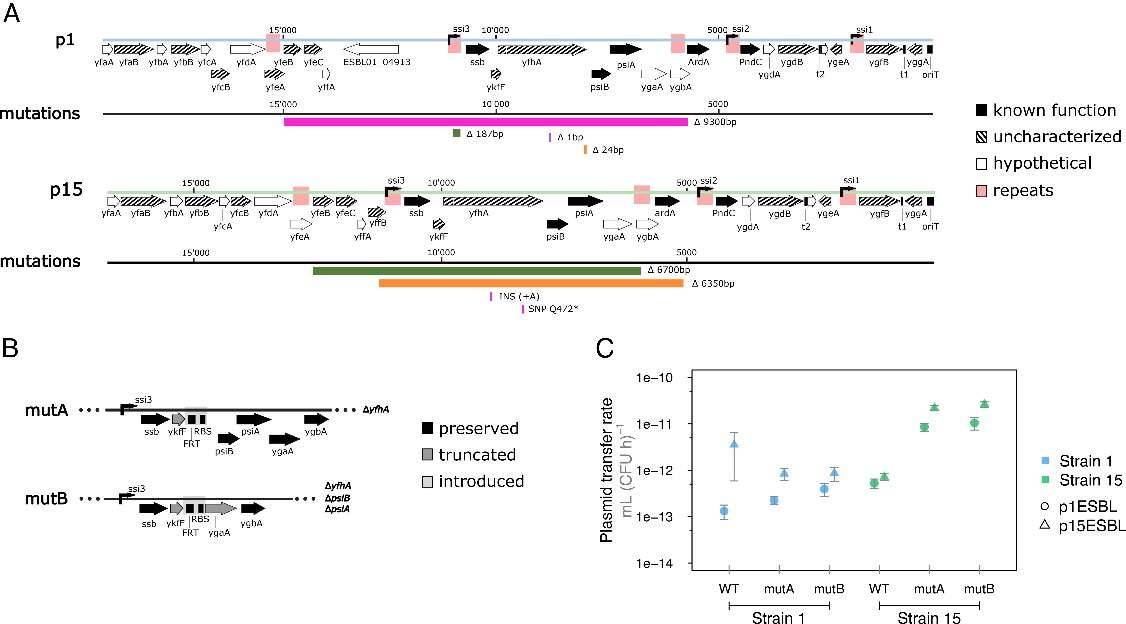 Fig 2. Genetic changes in the plasmid leading region increase transfer rate in a host-specific way.
Fig 2. Genetic changes in the plasmid leading region increase transfer rate in a host-specific way.
Conclusions
The authors found that plasmid persistence is driven more by rapid, strain-specific evolutionary changes than by initial stability traits. Mutations in the plasmid leading region, particularly in the ssi3 operon, affect transfer rates differently across strains. Their study highlights the importance of considering rapid evolution in managing successful bacterium-plasmid combinations and suggests further research to refine predictions in clinical settings.




 Fig 1. Genetic changes in chromosomes and ESBL plasmids after 15 d of serial passage without antibiotics.
Fig 1. Genetic changes in chromosomes and ESBL plasmids after 15 d of serial passage without antibiotics. Fig 2. Genetic changes in the plasmid leading region increase transfer rate in a host-specific way.
Fig 2. Genetic changes in the plasmid leading region increase transfer rate in a host-specific way.