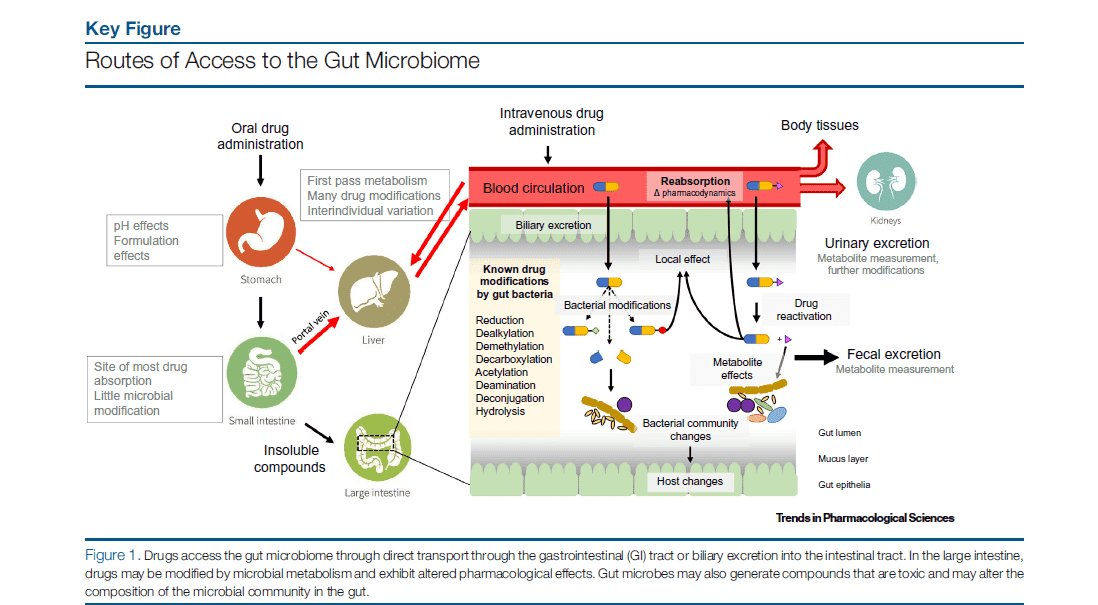
Introduction to Gut Microbiota
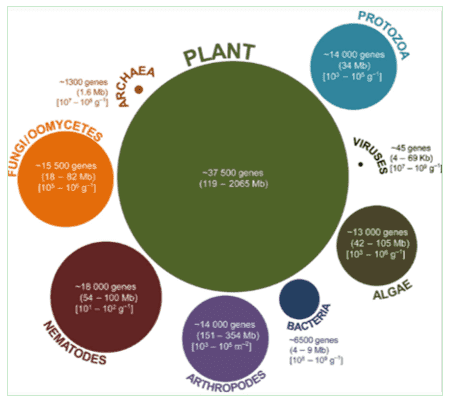
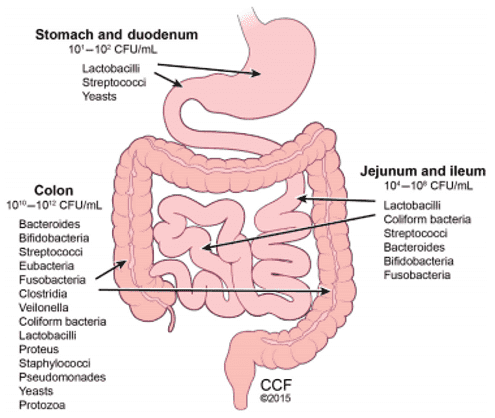
The gut microbiota represents a thriving and intricate community of symbiotic microbes inhabiting the animal gastrointestinal tract. Comparable to an organ in its own right, gut microbiota conduct a plethora of physiological functions that encompass metabolism, immunological regulation, and endocrine operations. Beneficial microbes play a cardinal role in normal gut functionality: they aid in moisture absorption, vitamin synthesis, the mitigation of harmful substances, and the prevention of pathogenic invasion. In contrast, an excess of harmful microbes can lead to invasive pathogens, alterations in gut motility, production of detrimental substances, and poor waste excretion.
The gut microbiota and its metabolic products have been identified as significant players in human health and disease; hence, studying the correlations between gut microbiota metabolites and diseases holds considerable potential for disease prevention and management.
CD Genomics proffers leading-edge, integrated research solutions for the study of gut microbiota, employing advanced metabolomics and high-throughput sequencing technologies.
Our Advantages:
- Detection of low abundance species of microbial communities
- Flexible sequencing strategies and data analysis
- Reliable, high-throughput and sensitive platform for gut metabolomics analysis
- Years of commercial experience in life sciences
- Quick and affordable services
The advancement in modern technology has fostered a shift towards an integrative approach in the study of gut microbiota. More researchers are harnessing the synergy of microbial sequencing and metabolomics analyses to comprehensively research the gut microbiota and its impact on human health.
Why Adopt Gut Microbiota Sequencing
Employing the advanced high-throughput sequencing technology of 16S rRNA, we can probe the diversity of microbes present in intestinal content or fecal samples. This approach not only permits the taxonomy of gut microbiota at the genetic level but also discloses core details such as microbial species richness, abundance, and distribution. Gathering comprehensive data on the microbiome will offer a wellspring of resources for intricate phenotype analysis, thereby expediting the discovery of therapeutic agents.
Our Gut Microbiota Sequencing Service
To explore the assembly of gut microbial communities, CD Genomics is equipped with various sequencing platforms which include Illumina, PacBio SMRT system and Nanopore system.
We have undertaken testing and analysis of a range of gut microbiota specimens, enabling us to assist in identifying the diversity and relative abundance of microbes resident in the gut, along with their phylogenetic relationships. This, thus, provides comprehensive insights into the gut microbiome at the genetic level.
Bioinformatics Analysis
| Basic Analysis | Routine Analysis (According to Customer Requirements) | Advanced Data Analysis |
| Sequence Filtering and Trimming | Heatmap | Phylogenetic Tree |
| Sequence Length Distribution | VENN | LDA-Effect Size (LEfSe) |
| OTU Clustering and Species Annotation | Principal Components Analysis (PCA) | Network Analysis |
| Diversity Index | Microbial Community Structure Analysis | Correlation Analysis |
| Shannon-Wiener Curve | α Diversity Index Analysis | |
| Rank-Abundance Curve | Matastats Analysis | |
| Rarefraction Curve | Weighted Unifrac test | |
| Multiple Contrast | CCA/RDA Analysis | |
| Heatmap | ||
| Principal Components Analysis (PCA) |
Workflow


Sample Requiremen
- DNA sample: total DNA ≥ 300 ng, concentration ≥ 10 ng/uL, OD260/280=1.8-2.0
Be ensure that the DNA is intact and non-degradative, avoiding repeated freezing and thawing cycles.
Please use enough dry ice or ice packs during shipment.
We provide a broad range of oral and fecal sample collection kits for microbial recovery and analysis.
Deliverables
- Experimental procedure
- Experiment parameter
- Original data
- Results of sequencing analysis
Sequencing Analysis of Gut Microbiota FAQ:
- 1. What is gut microbiota responsible for?
- 2. What are the 3 types of gut microbiota?
- 3. What is the role of 16S rRNA sequencing analysis in the study of gut microbiota?
- 4. What are the methods for gut microbiome sequencing?
Demo:
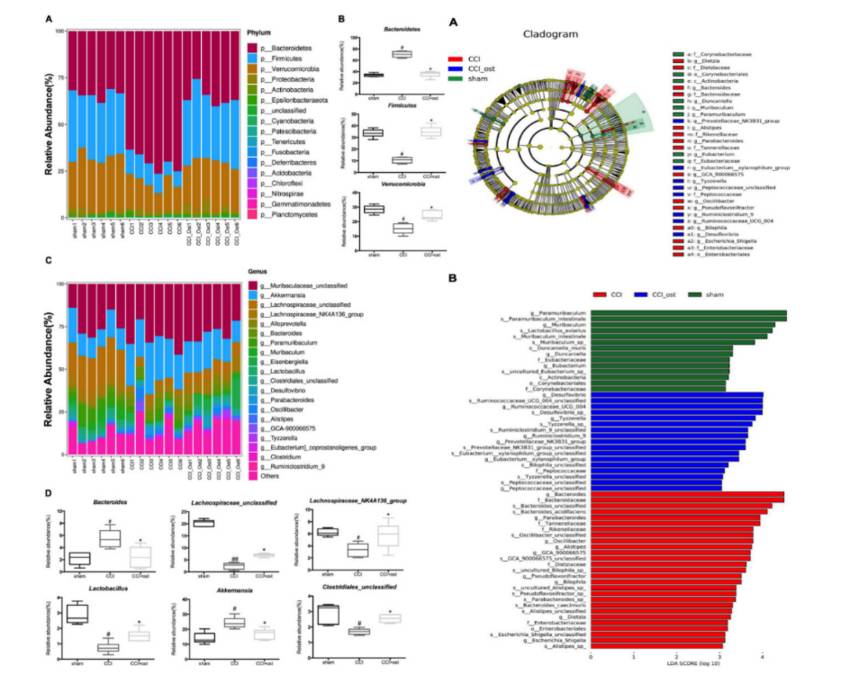 Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice. (Ruili Li et al., 2022)
Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice. (Ruili Li et al., 2022)
Introduction to Gut Microbiota Metabolomics
Gut microbiota primarily interacts with its host through the intimate exchange of information via their small molecule metabolites. This interaction plays a pivotal role in various physiological aspects such as metabolism, immunity, diseases, as well as the development and regulation of the nervous system. These microbes significantly influence the host's digestive absorption capacity, resistance to infection, and the host's response to therapeutic drugs, thereby dictating health and disease status in the organism.
Microbiome metabolomics, a field that employs metabolomics technologies to analyze the dynamic variations in the metabolites of gut microbes, aims to identify key microbial metabolites associated with pathological variations in the host. By determining biologically relevant biomarkers associated with specific diseases, microbiome metabolomics can provide innovative methodologies for clinical diagnoses, while charting new directions and insights for therapeutic developments.
Our Gut Metabolomics Services
In a bid to decipher the complexities of the gut microbiome, CD Genomics has established an accurate, sensitive and high-throughput analytics platform for gut metabolomics. Different metabolomics methodologies are employed depending on the design and requirements:
- Untargeted metabolomics analysis of the gut microbiota metabolites
- Targeted metabolomics analysis of the gut microbiota metabolites
- Lipidomics analysis of the gut microbiota metabolites
- Stable isotope-assisted metabolic flux analysis of the gut microbiota metabolites
The metabolites we are capable of analyzing encompass short-chain fatty acids, bile acids, secondary bile acids, metabolites of choline, phenolic compounds, indoles, polyamines, lipids, vitamins, hormones, lipopolysaccharides, and trimethylamine, among others.
Bioinformatics Analysis
- Univariate statistical analysis
- Multivariate statistical analysis
- Variance analysis
- Annotation analysis of differential substances
- Metabolic pathway analysis
Gut Microbiota Metabolomics Workflow
Sample Requirement
- Serum, plasma ≥ 200 ul/sample
- Urine ≥ 500 ul/sample
- Tissue ≥ 200 mg/sample
- Cell ≥ 1×107/sample
- Cerebrospinal fluid ≥ 200 ul/sample
- Stool and intestinal contents ≥ 500 mg/sample
- For other sample types, please consult our technicians.
Note: Avoid repeated freezing and thawing.
Deliverables
- Experimental procedure
- Mass spectrometry parameters
- Mass spectrum picture
- Original data
- Results of metabolomics analysis
Gut Microbiota Metabolomics FAQ
- 1. What are the metabolites from the gut microbiota?
- 2. What problem is metabolomics used to solve?
- 3. What are the methods of metabolomic analysis?
Demo:
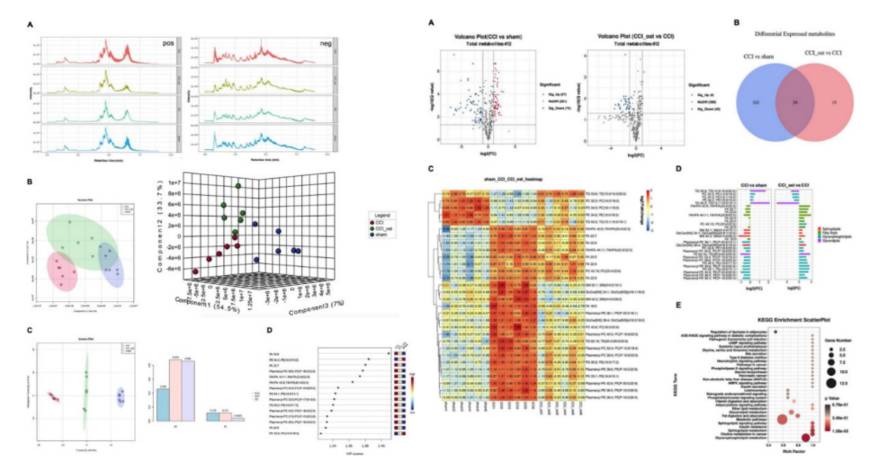 Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice. (Ruili Li et al., 2022)
Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice. (Ruili Li et al., 2022)
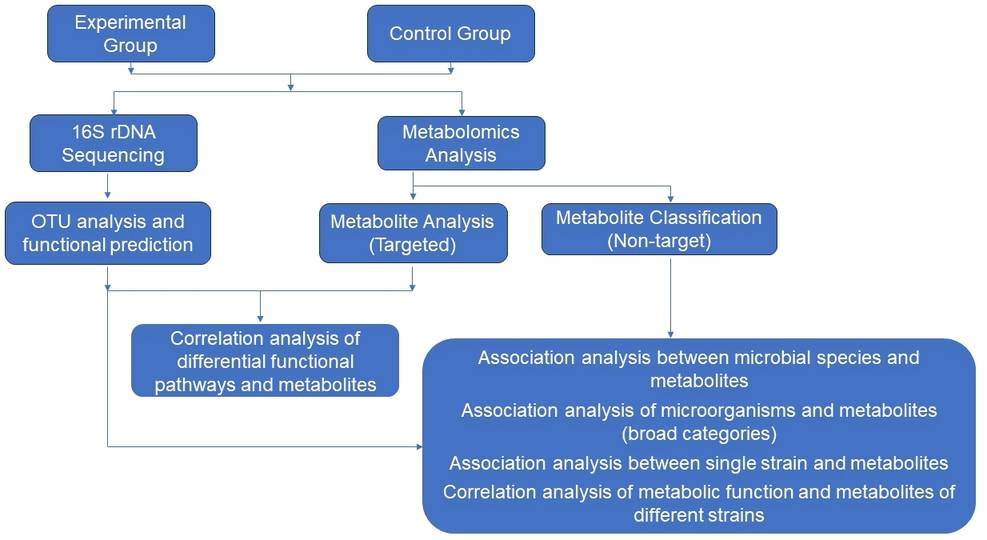
The Implications of Integrative Analysis Through Metabolomics and Sequencing
By integrating and interpreting 16S rRNA sequencing data and metabolomics, it is possible to uncover associations between specific microbial constituents and metabolite groups. This can aid in selecting key strains attributable to variations in microbial metabolic status or functional metabolites. Additionally, exploring the interconnection between metabolites, microbes, and disease through combined metabolomic and 16S rRNA sequencing analyses may facilitate the discovery of biomarkers consequential to disease, fostering their further application in clinical diagnostics.
Our Integrative Analysis Services
CD Genomics, harnessing a varied array of analytical tools such as GC-MS, LC-MS, and Nuclear Magnetic Resonance (NMR), provides targeted and non-targeted metabolomic analysis services for a broad range of samples. Along with 16S rRNA sequencing service integrated with customizable bioinformatics, CD Genomics offers a holistic one-stop solution from experiment design, sample testing, to data analysis– catered to meet multiple detection needs.
Bioinformatics Analysis
- Diversity Analysis
- Functional analysis
- Correlation analysis
- Metabolic pathway analysis
- Annotation analysis of differential substances
- Association analysis between microbial species and metabolites
- Correlation analysis of differential functional pathways and metabolites
- Correlation analysis of metabolic function and metabolites of different strains
- Association analysis of microorganisms and metabolites (broad categories)
Gut Microbiota Integrative Analysis Workflow
Sample Requirement
- DNA sample: total DNA ≥ 300 ng, concentration ≥ 10 ng/uL, OD260/280=1.8-2.0
Be ensure that the DNA is intact and non-degradative, avoiding repeated freezing and thawing cycles.
Please use enough dry ice or ice packs during shipment.
We provide a broad range of oral and fecal sample collection kits for microbial recovery and analysis. - Serum, plasma ≥ 200 ul/sample
- Urine ≥ 500 ul/sample
- Tissue ≥ 200 mg/sample
- Cell ≥ 1×107/sample
- Cerebrospinal fluid ≥ 200 ul/sample
- Stool and intestinal contents ≥ 500 mg/sample
- For other sample types, please consult our technicians.
Note: Avoid repeated freezing and thawing.
Analysis of genomics and metabolomics correlations requires that identical types of samples are separately prepared prior to cryopreservation for both microbial sequencing and metabolomics profiling. This preparation involves taking samples of different types during the same time window, a process without which association would be impossible.
Deliverables
- Experimental procedure
- Experiment parameter
- Mass spectrum picture
- Original data
- Results of metabolomics analysis and microbiome sequencing
Integrative Analysis Through Metabolomics and Sequencing FAQ
- 1. What are the advantages of integrating metabolomics and genome analysis?
- 2. Where integration studies of metabolomics and genomic data can be applied?
- 3. What is multi-omics integrative analysis?
Demo:
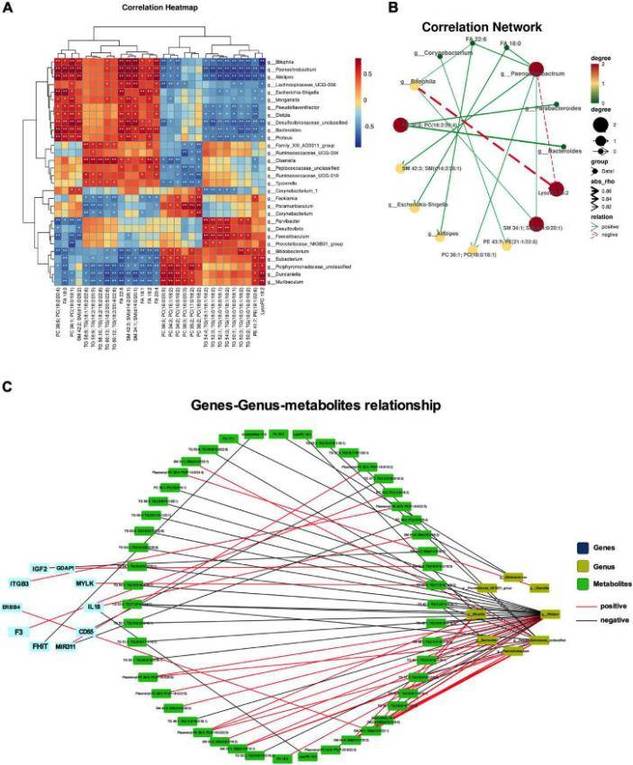
Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice. (Ruili Li et al., 2022)
Reference
- Li R, Wang F, Dang S, et al. Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice. Frontiers in Physiology, 2022, 13: 813626.