Background
Soil salinization affects crop productivity globally, reducing yields and increasing soil-borne pathogens. Intercropping, such as peanut and sorghum, can improve resource use and stress tolerance. This study uses 16S rDNA and ITS sequencing to analyze bacterial and fungal communities in peanut-sorghum intercropping systems under saline conditions, aiming to enhance soil stability and optimize cropping systems.
Materials & Methods
Method:
-
Full-Length 16S rDNA genes Sequencing
- Full-Length ITS Sequencing
Data Analysis:
-
Annotation
- Microbial diversity analysis
- PCoA analysis
- Linear discriminant analysis (LDA)
- Effect size measurements (LEfSe) analysis
Results
OTU Analysis: Sequencing depth was adequate, revealing 1,531 bacterial and 284 fungal OTUs with high coverage. Venn diagrams showed common OTUs across samples, with reduced overlap under salt stress.
Community Composition: Salt treatment altered bacterial and fungal communities, increasing Bacteroidota and Ascomycota while decreasing Acidobacteriota and Mucoromycota.
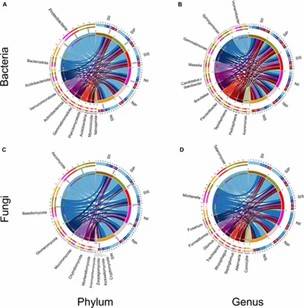 Fig 1. Relative abundance of the peanut rhizosphere (IP), sorghum rhizosphere (IS), and interspecific interaction zone (II) microbial communities at the phylum and genus levels under different soil conditions.
Fig 1. Relative abundance of the peanut rhizosphere (IP), sorghum rhizosphere (IS), and interspecific interaction zone (II) microbial communities at the phylum and genus levels under different soil conditions.
Beta Diversity: Distinct bacterial and fungal community structures were observed between salt-treated and normal soils, with different clustering patterns under varying conditions.
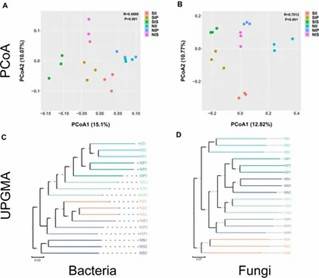 Fig 2. Principal coordinate analysis and unweighted pair group method with arithmetic mean analysis.
Fig 2. Principal coordinate analysis and unweighted pair group method with arithmetic mean analysis.
Microbial Taxa Abundance: Core bacterial taxa shifted under salt stress, with increased Bacteroidetes and Verrucomicrobiota. Fungal communities showed significant changes, especially in Ascomycota and Mucoromycota, with salt treatment.
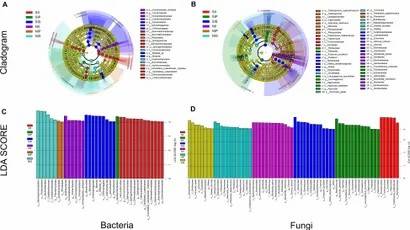 Fig 3. Linear discriminant analysis effect size.
Fig 3. Linear discriminant analysis effect size.
Conclusions
Soil salinity significantly shapes microbial communities, with peanut driving changes more than sorghum. While fungal communities in different soil conditions shared main phyla, their taxonomic compositions differed. Under salt stress, interspecies interactions promote specific microbial recruitment. Future research should clarify these mechanisms and their applicability to natural ecosystems.

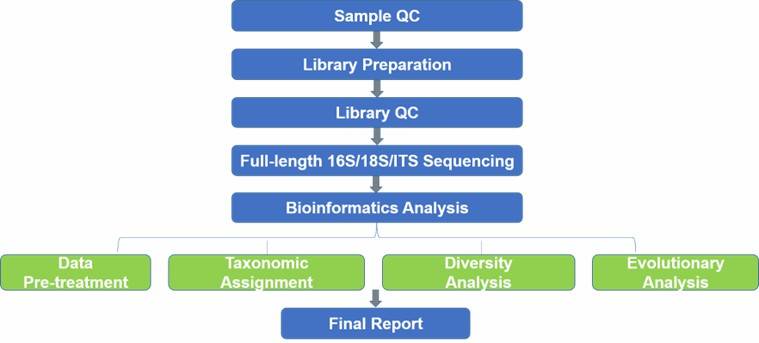
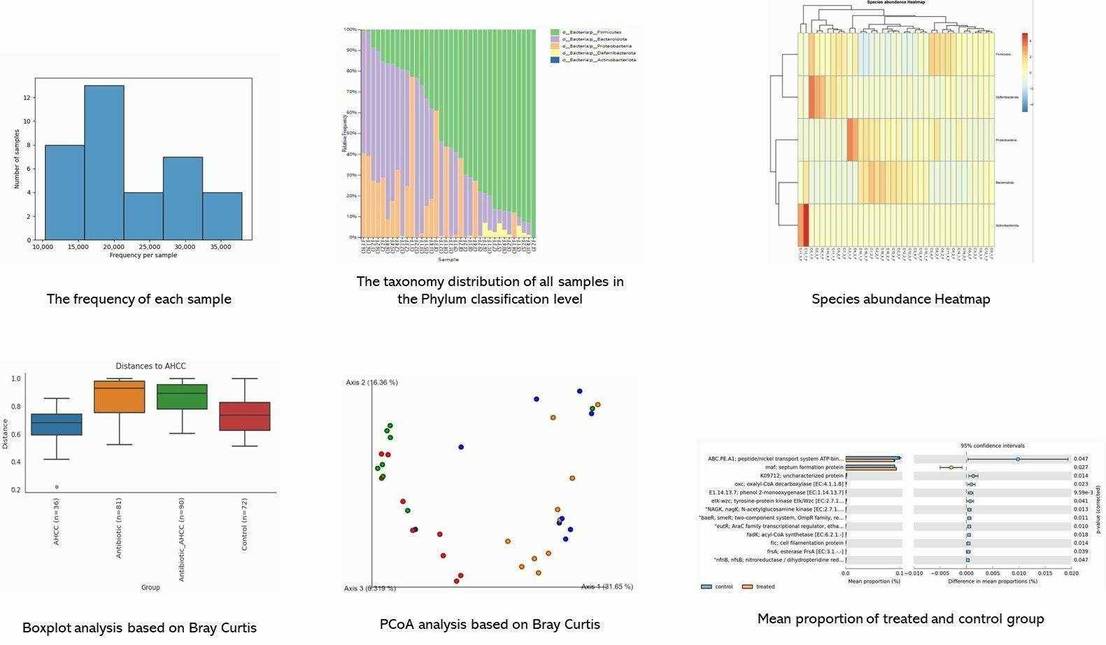

 Fig 1. Relative abundance of the peanut rhizosphere (IP), sorghum rhizosphere (IS), and interspecific interaction zone (II) microbial communities at the phylum and genus levels under different soil conditions.
Fig 1. Relative abundance of the peanut rhizosphere (IP), sorghum rhizosphere (IS), and interspecific interaction zone (II) microbial communities at the phylum and genus levels under different soil conditions. Fig 2. Principal coordinate analysis and unweighted pair group method with arithmetic mean analysis.
Fig 2. Principal coordinate analysis and unweighted pair group method with arithmetic mean analysis. Fig 3. Linear discriminant analysis effect size.
Fig 3. Linear discriminant analysis effect size.









