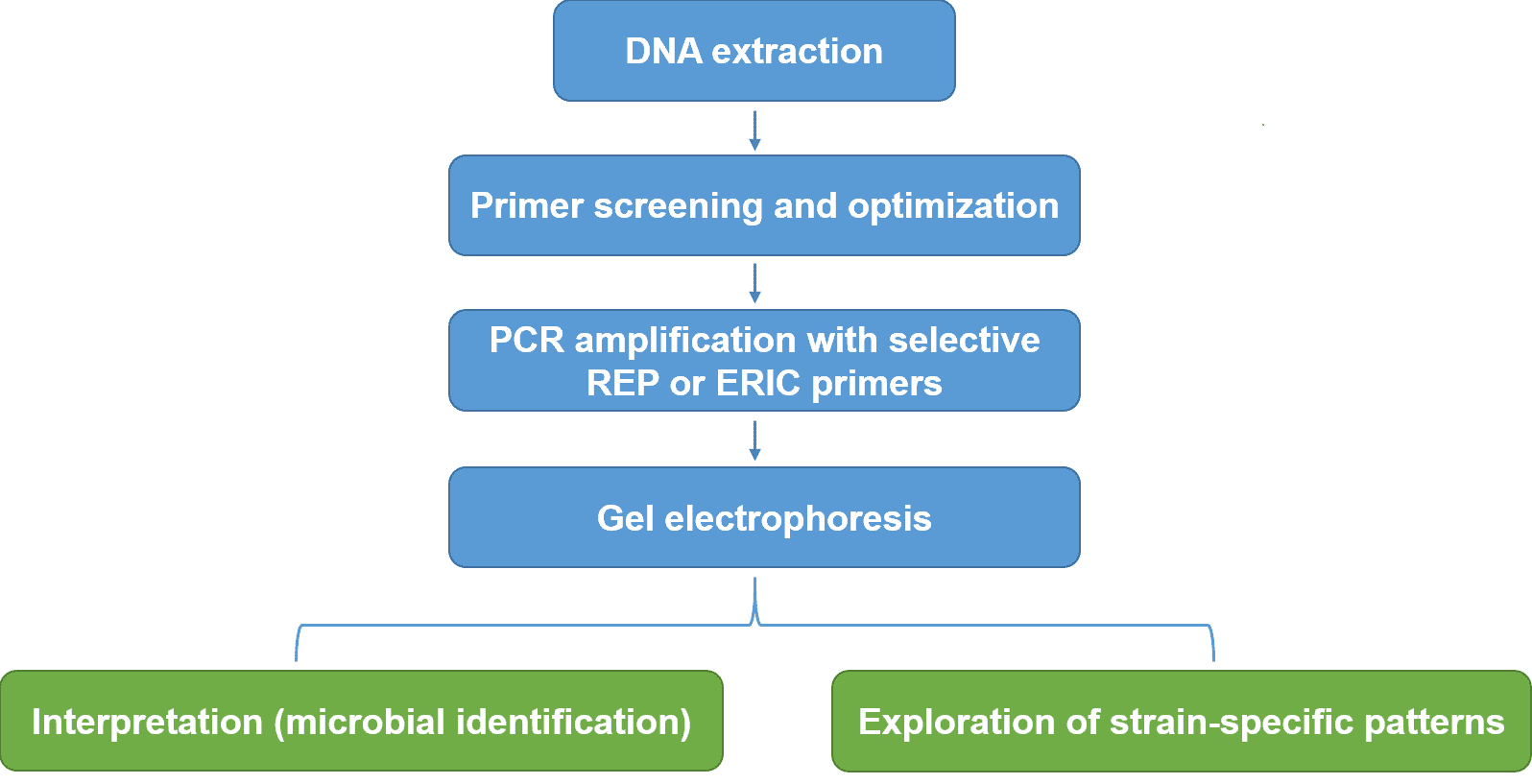
Rep-PCR profiling service workflow
Sample Requirement
-

- 1.8 < OD260/280 < 2.0, no degradation or contamination.
- Mixed microbial genomic DNA: total amount > 1 ug, concentration > 10 ng/ul.
- Gram-positive bacteria are not easy to extract from a complex microbial community due to the relatively thick cell wall, and the use of conventional methods may result in the loss of strains. Please provide us with more than 10 ml of microbial solution (OD600 > 1.5) or the microbial pellet.
Sampling kits: We provide a complete range of microbial sampling kits for clients, including microbial collection products, DNA/RNA isolation kits, and accessories for storage and mailing.
Deliverables: The experimental report includes information on experimental methods, primers, PCR conditions, gel electropherogram, etc.
Reference
- Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. Journal of clinical microbiology, 1999, 37(6): 1661-1669.