The Importance of Microbes in the Air
Microorganisms such as bacteria, virus, and fungi am teeming in the air. These biological particles contributes to the ecological balance and play an important role in agriculture, the biosphere, cloud formation, global climate, and atmospheric dynamics. However, they also affect the environmental equality, and even act as a medium for the spread that leads to air pollution and human diseases. Airborne microorganisms are a major cause of human respiratory diseases, such as asthma, allergies, and pathogenic infections. Both cultivation-dependent and –independent methods can be used to explore microbial diversity in the air, uncovering the potential impacts on the natural environment and human disease.
Accelerate Your Microbial Research in the Air
We analyze the microbiome of various environments including indoor and outdoor air, and their impact on human health using sequencing techniques like metagenomics sequencing, 16S/18S/ITS sequencing, and metranscriptomic sequencing. RT-qPCR is also used for achieving the qualitative and quantitative analysis of DNA or RNA. Bioinformatics analysis is required to process raw reads and determine the taxonomic classifications, biological functions for ecological environment, and effects on human health.
The Main Research Directions
- The sources of microorganisms and the relative contribution of each.
- The community structure and abundance of airborne microorganisms at different places.
- The relationship between airborne microorganisms and pollution of antibiotic resistance gene.
- The airborne microorganisms and respiratory diseases of humans.
What Can We Do?
- 1. Measuring the types and abundance of microorganisms in the samples;
- 2. Qualitative and quantitative analysis of certain microorganisms in the samples;
- 3. Obtaining the microorganism strain by anaerobic culture.
Note: Our service is for research use only, and not for therapeutic or diagnostic use.
Detectable Objects
The various microbiome samples from indoor or outdoor environments. We have ever analyzed various samples collected from hospitals, schools, industrial buildings, etc.
Detectable Microorganisms
Bacteria, fungi, virus.
Detection Methods
Next-generation sequencing (NGS), long-read sequencing, PCR-DGGE, Real-time qPCR, anaerobic culture.
Technical Platforms
Illumina HiSeq/MiSeq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE, Real-time qPCR, anaerobic culture.
Sample Requirement
-

- DNA samples: total DNA ≥ 300 ng, concentration ≥10 ng/μL, OD260/280 = 1.8-2.0.
- Non-degraded DNA. Avoid repeated freezing and thawing. Samples should be transported with ice packs or dry ice.
Workflow
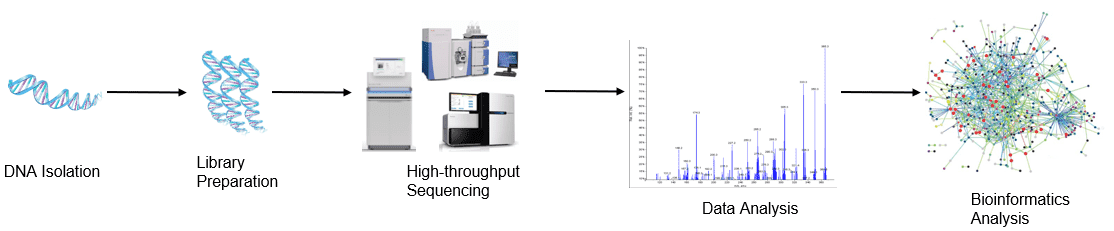 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
| OTU Clustering | Distribution-Based OTU-Calling |
| Rarefaction Curve | |
| Shannon-Wiener Curve | |
| Rank Abundance Curve | |
| Diversity Index | |
| OTU-Based Analysis | Heatmap |
| VENN | |
| Principal Components Analysis (PCA) | |
| Hierarchical Clustering | |
| Comparative Analysis | RDA/CCA |
| PCA/PCoA | |
| Network Analysis | |
| Functional analysis | BLASTX, KEGG, eggNOG, CAZy, CARD, ARDB etc. |
| PICRUSt | |
| TAX4Fun | |
| Phylogenetic Analysis | (Un)Weighted Unifrac |
| Phylogenetic Trees |
Reference
- Liu Z, et al. Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environmental International, 2019, 129, 208-220.
- H.K. Park, et al. Bacterial diversity in the indoor air of pharmaceutical environment. Journal of Applied Microbiology, 2013, 116, 718-727.
- Wu S, et al. Microbial community structure and distribution in the air of a powdered infant formula factory based on cultivation and high-throughput sequence methods. Journal of Dairy Science, 2018, 101, 1-12.