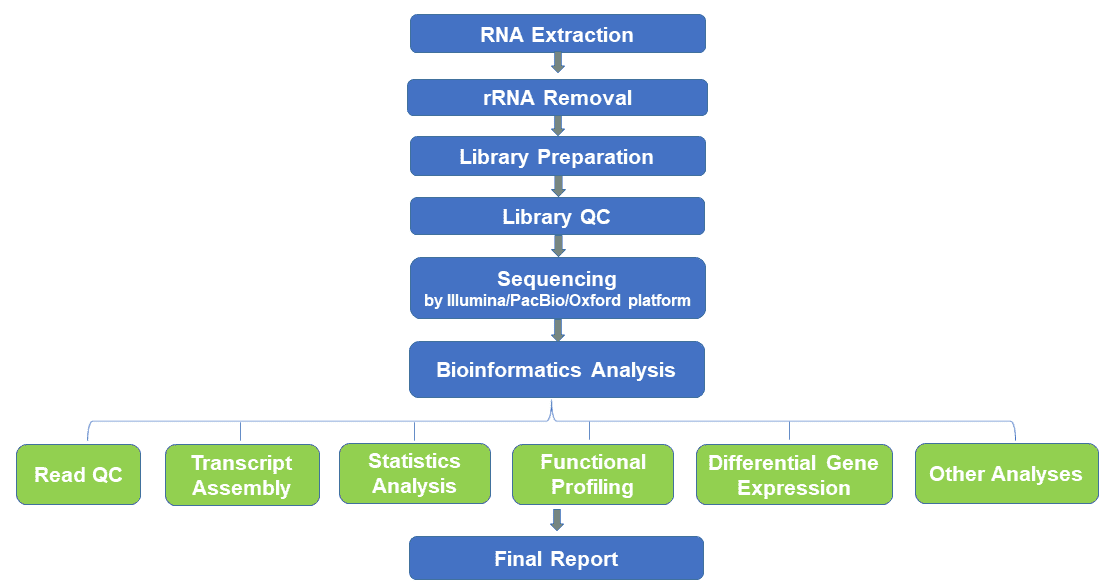
Eukaryotic RNA sequencing, based on high-throughput sequencing, refers to the study of all transcripts of specific tissues or cells of eukaryotic organisms. It serves as a powerful tool in understanding the complexity of the eukaryotic transcriptome. Depending on the availability of reference transcriptomes/genomes, eukaryotic RNA-seq can be divided into reference-based transcriptome assembly and de novo transcriptome assembly. Here, we have established standardized eukaryotic RNA-seq platforms based on NGS, PacBio SMRT, and Oxford nanopore instruments to identify common and novel transcripts in microbial eukaryotes such as fungi and yeast, without bias. PacBio SMRT sequencing and Oxford nanopore sequencing can provide full-length transcript sequences, and also detect more transcript overlaps, polycistronic RNAs, splice and transcript variants.
Eukaryotic RNA-seq detects both unknown and rare transcripts, provides quantitative analysis of transcripts, reveals differences in gene expression levels of different samples, and performs structural analysis for discovering variable splicing sites, gene fusions, SNPs and InDel sites. Eukaryotic RNA sequencing is important for revealing the regulation of gene expression in eukaryotic cells, exploring host-pathogen interaction, and monitor disease progression.