The Importance of Detecting Blood Microbiome
A large part of bacterial, archaeal, viral, and fungal microbial taxa consists of the human microbiome. While many of these microorganisms are commensal, some are harmful to humans and many are symbiotic. Our livelihoods are strongly intertwined with the microbes we allocate our bodies with, despite whether their appearance is advantageous, insignificant, or harmful. Over the past few years, human microbiome research has grown quickly.
Blood has always been considered free of microorganisms. But recent DNA sequencing methods have shown that 1 mL of blood contains about 1,000 bacterial cells. These bacteria are usually dormant. The profiling and characteristics of the blood microbiome by methods such as next-generation sequencing and long-read sequencing can facilitate the research on many diseases including bacterial infections and even cancer.
Accelerate Research and Practice in Blood Microbiome
Based on our next-generation sequencing (NGS), PacBio SMRT sequencing, and Nanopore sequencing platforms, we have tested and analyzed various blood microbiome samples. We can help you identify the microorganisms presented in the blood vessel, their relative abundance, and evolutionary relationships. We also study the correlation between blood microbes and human health, the function of blood microorganisms from gene level can also be studied, all of which are of great importance for scientific research, the development of supplements and therapies.
The Main Research Direction of Blood Microbiome
- Explore differences in microbial composition and content between disease and healthy groups
- To study and compare the differences in blood flora structure between individuals at different stages of intervention
- To study the changes in blood flora in the same individual at different stages of development
- To study the changes in blood flora during the development of a disease in order to find biomarkers associated with disease
What Can We Do?
- 1. Identification and discovery of blood microorganisms
- 2. Quantitation of microorganisms in the blood vessel
- 3. Evolutionary and functional analysis of microorganisms in the blood vessel
- 4. Relationship between microorganisms and diseases
Note: Our services are for research use only, not for disease diagnosis and treatment.
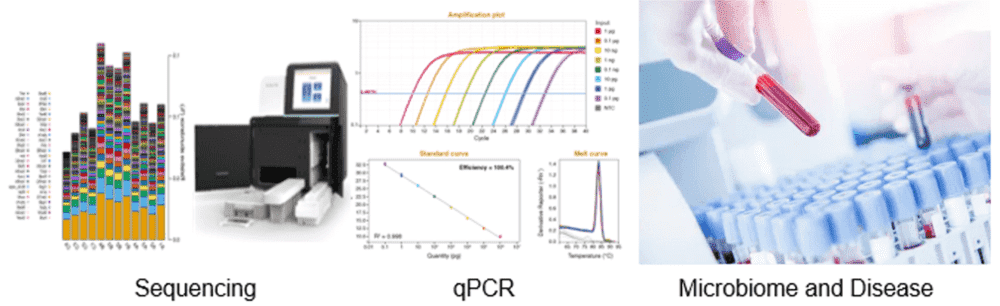
Detectable Objects
We have successfully analyzed reproductive blood microbiome samples from human and other animals.
Detectable Microorganisms
Fungi, bacteria, and viruses, etc.
Technical Platforms
We are equipped with Illumina HiSeq/MiSeq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time qPCR, clone library, and other detection platforms.
Sample Requirement

DNA sample: DNA ≥ 500ng, concentration ≥ 10ng/ul, and OD260/280 = 1.8-2.0.
Ensure the DNA is not degraded or slightly degraded and avoid repeated freezing and thawing cycles.
Please use enough ice packs or dry ice during transportation.
Workflow
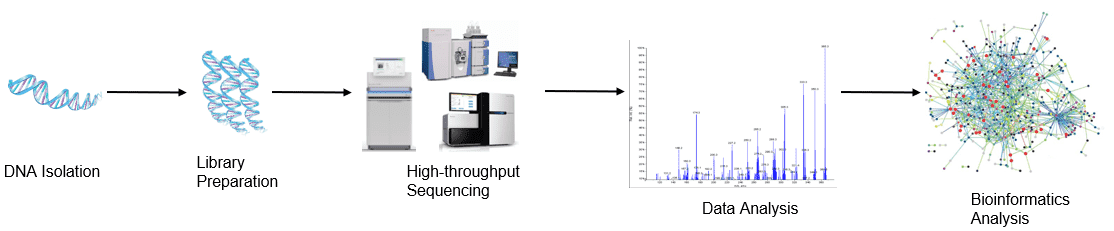 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
| OTU Clustering | Distribution-Based OTU-Calling |
| Rarefaction Curve | |
| Shannon-Wiener Curve | |
| Phylogenetic Tree | |
| Rank Abundance Curve | |
| Relative Abundance | Heatmap |
| VENN | |
| Principal Components Analysis (PCA) | |
| Hierarchical Clustering | |
| Community Structure | RDA/CCA Analysis |
| Multiple Contrast | |
| Network Analysis | |
| Functional analysis | BLASTX, KEGG, eggNOG, CAZy, CARD, ARDB etc. |
| PICRUSt | |
| TAX4Fun | |
| Custom Analysis | Custom Analysis |