Background
Bacterial communities in aquatic systems are vital for nutrient cycling and energy flow. They affect organic matter decomposition and element cycling at the sediment-water interface. Freshwater sediments, with high microbial biomass and diversity, play a key role in nutrient transformation. This study used 16S rRNA gene sequencing and metagenomic sequencing to analyze microbial diversity and nutrient dynamics in Lake Caviahue, focusing on bacterial roles in phosphorus cycling and community variations across different lake strata.
Materials & Methods
Sample preparation:
-
Water
- Sediments
- DNA extraction
Data Analysis:
-
Sequence quality and processing
- De novo assembly
- Diversity analysis
Results
In acidic Lake Caviahue, microbial biomass and diversity are higher in the bottom strata compared to the upper layers. The average microbial abundance was significantly greater in the metalimnion and bottom layer than in the epilimnion. Using targeted metagenomic analysis, it was found that microbial diversity increases from the top to the bottom strata. A microcosm bioassay showed that sediment and pore water bacteria contribute to nutrient retention and release, with significant differences in microbial community structure and diversity between test and control columns. Despite initial differences in microbial load, both columns showed increased microbial growth and similar trends in nutrient utilization over time.
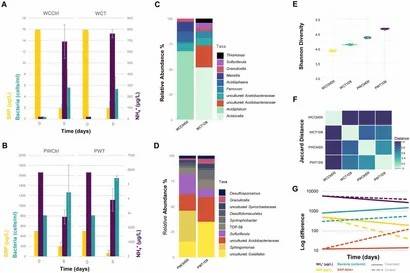 Figure 1. Contribution of sediment and pore water bacterial community to nutrient recirculation of Lake Caviahue.
Figure 1. Contribution of sediment and pore water bacterial community to nutrient recirculation of Lake Caviahue.
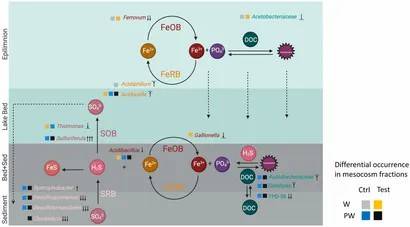 Figure 2. Conceptual model of the biogeochemical cycling of nutrients, sulfur, and iron at the sediment-water interphase of Lake Caviahue.
Figure 2. Conceptual model of the biogeochemical cycling of nutrients, sulfur, and iron at the sediment-water interphase of Lake Caviahue.
Conclusions
In Lake Caviahue, bacterial abundance and diversity are highest in the bottom strata and peak in autumn. Seasonal changes affect pH, temperature, and nutrient levels. Both bacteria and algae influence nutrient cycling, with a significant syntrophic relationship between them. Sediment microorganisms play a key role in nutrient utilization and mobilization.




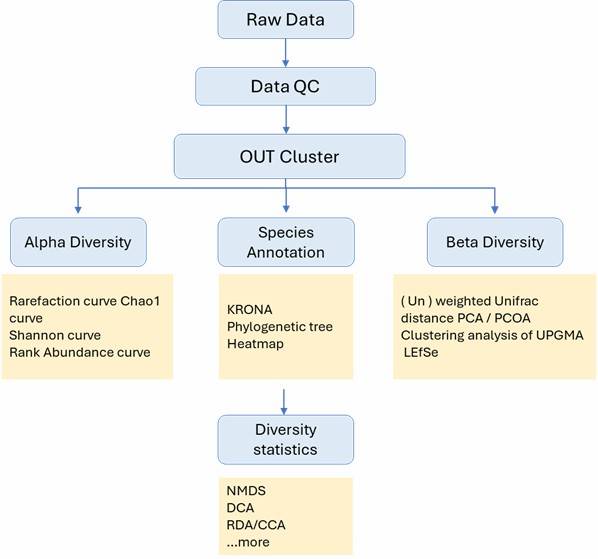
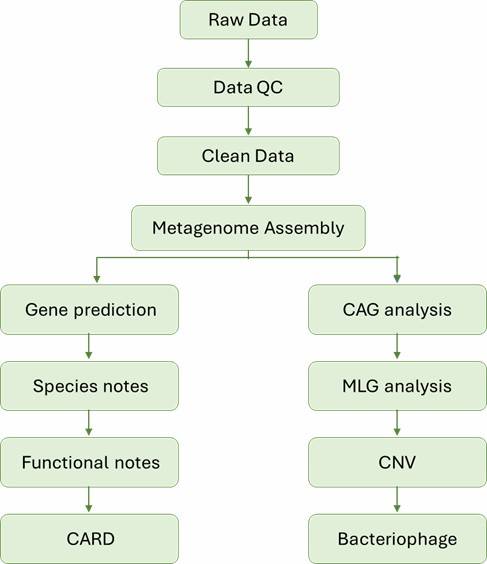
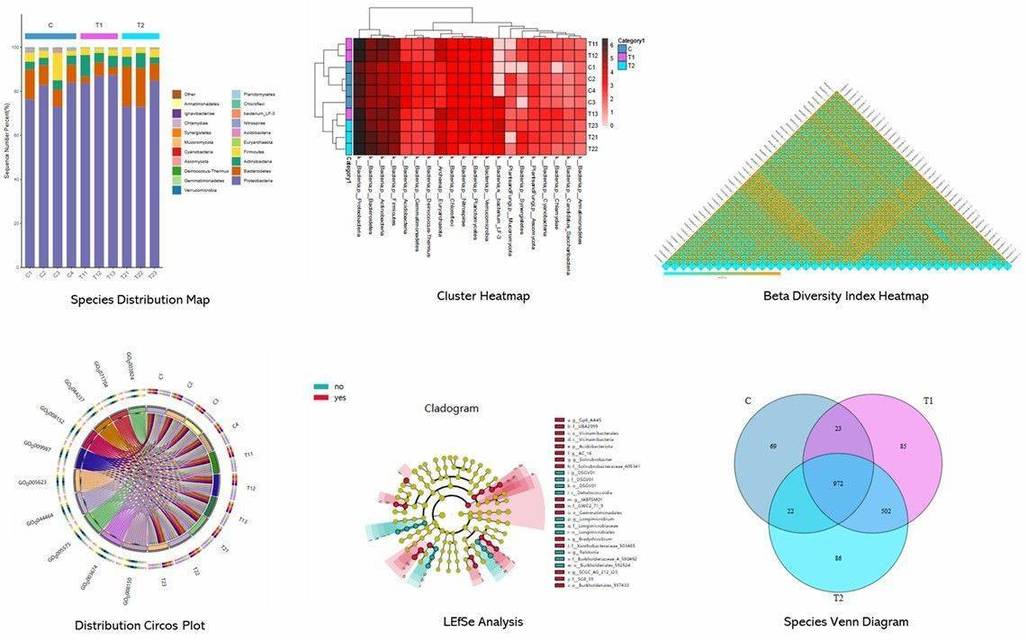

 Figure 1. Contribution of sediment and pore water bacterial community to nutrient recirculation of Lake Caviahue.
Figure 1. Contribution of sediment and pore water bacterial community to nutrient recirculation of Lake Caviahue. Figure 2. Conceptual model of the biogeochemical cycling of nutrients, sulfur, and iron at the sediment-water interphase of Lake Caviahue.
Figure 2. Conceptual model of the biogeochemical cycling of nutrients, sulfur, and iron at the sediment-water interphase of Lake Caviahue.