Our comprehensive microbial identification platform, encompassing methodologies such as Rep-PCR, MicroSEQ®, NGS-based microbial identification, multi-locus sequence typing, and mycoplasma detection, is designed to identify microbial species with efficiency and cost-effectiveness. This system supports high-throughput and streamlined processes to deliver data with exceptional accuracy and sensitivity, thereby advancing your microbial and laboratory research objectives.
Our Advantages:
- High Accuracy: We achieve high accuracy in microbial identification by integrating both phenotypic and genotypic methods to minimize misidentification.
- Rapid Turnaround: Our use of advanced sequencing technologies enables rapid processing, delivering results in a timely manner.
- Comprehensive Analysis: Our detailed reports include species identification, phylogenetic relationships, and potential health implications.
- Custom Solutions: We tailor our microbial identification services to meet specific client needs, accommodating various sample types and identification objectives.
What is Microbiological Identification
Microbiological identification entails a spectrum of methodologies aimed at discerning the identity of microorganisms by examining their distinct physical, biochemical, and genetic profiles. These minute entities, despite their microscopic dimensions, serve pivotal roles in ecosystems by significantly contributing to the biodiversity of various environments such as soil, water, and air. Moreover, microorganisms profoundly influence human health; they participate in beneficial processes including digestion and immune regulation, while also being implicated in infections and diseases.
The process of microbial identification is instrumental in elucidating microbial diversity, identifying pathogenic organisms, and assessing treatment efficacy in clinical settings. Methodologies for identification predominantly fall into two categories: phenotypic identification, which emphasizes observable characteristics such as morphology and biochemical reactions, and genotypic identification, which utilizes molecular techniques to analyze genetic material, offering enhanced precision.
Methods of Microbial Identification
- Phenotypic Identification
Phenotypic identification involves methods that characterize microorganisms based on observable traits and biochemical properties. This includes techniques such as the API and VITEK systems, MALDI-TOF MS, and the Biolog system.
The API and VITEK systems are automated platforms that classify microorganisms by assessing their biochemical and physiological characteristics, streamlining the identification process through comparison with comprehensive databases; however, their accuracy may be compromised by the quality of these databases, particularly for certain species like Bacillus and cocci. Similarly, MALDI-TOF MS employs mass spectrometry to quickly and accurately analyze microbial protein profiles, making it ideal for high-throughput environments, though it depends on a robust database for reliable results, necessitating additional methods for ambiguous identifications. The Biolog system evaluates microbial metabolic activity based on their ability to utilize various carbon sources, providing a user-friendly interface and extensive metabolic profiles, but its effectiveness also hinges on the quality of the underlying database.
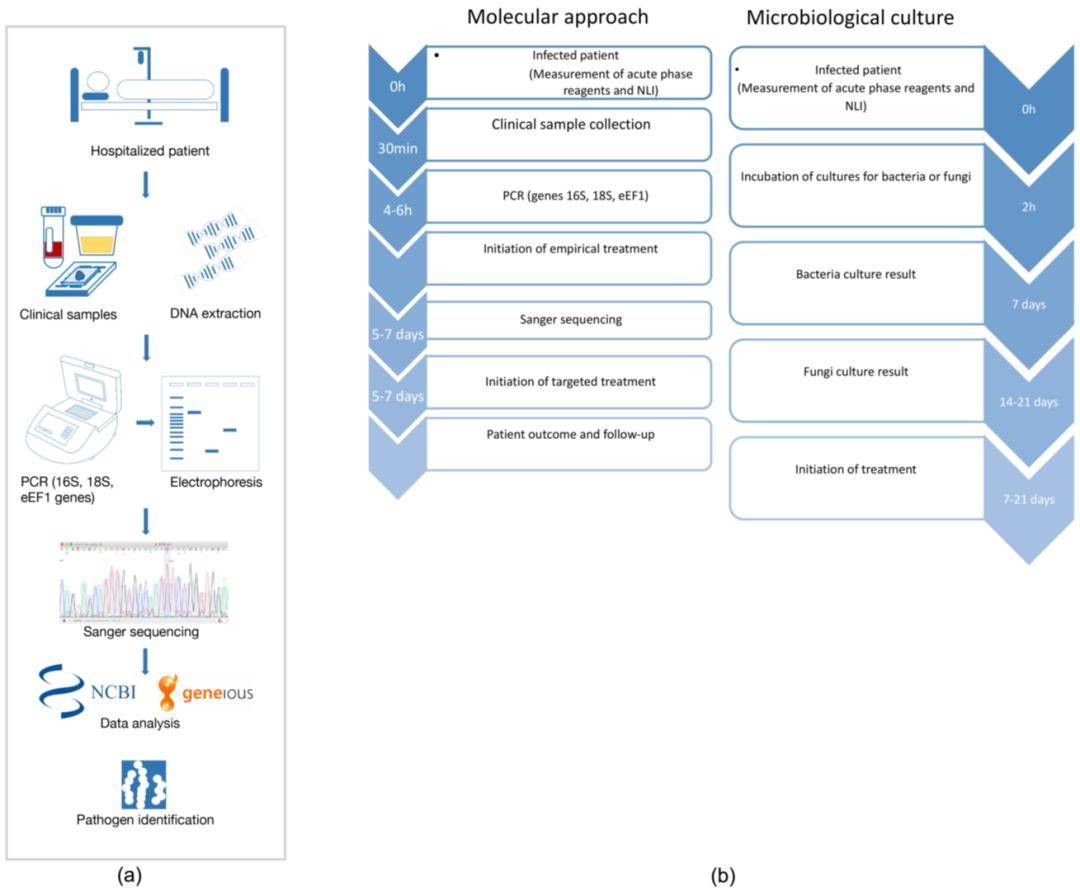
Genotypic identification methods utilize genetic information to accurately classify microorganisms, providing insights beyond observable traits. Key techniques in this category include Whole Genome Sequencing (WGS), 16S rRNA sequencing, genetic fingerprinting, and additional molecular techniques.
Whole Genome Sequencing (WGS) provides detailed insights into the genetic composition of microorganisms, facilitating accurate identification and traceability; however, it is more costly and resource-intensive than alternative methods. In contrast, 16S rRNA sequencing targets the conserved 16S rRNA gene found in all bacteria, enabling species identification based on genetic sequences, though it may lack resolution when differentiating closely related species. Genetic fingerprinting utilizes restriction enzymes to fragment microbial DNA, creating unique patterns for comparison with reference databases, yet it can struggle to distinguish closely related strains. Additionally, other genotypic methods, including quantitative PCR (qPCR), gene chips, and PFGE/DGGE, offer further identification options: qPCR quantifies specific DNA sequences, gene chips detect multiple genetic markers simultaneously, and PFGE/DGGE facilitate strain-level analysis through size or sequence variation.
Applications of Microbial Identification
The identification of microorganisms is critical for quality control, contamination detection, root cause analysis, and various microbiological studies. Consequently, microbial identification is extensively employed across diverse sectors, including agricultural production, environmental protection, research laboratories, academic institutions, biotechnology, and the pharmaceutical industry. Moreover, the precise identification of microbial pathogens is indispensable for understanding and managing numerous infectious disease syndromes.
Please feel free to reach out if you have any further inquiries or require additional information.


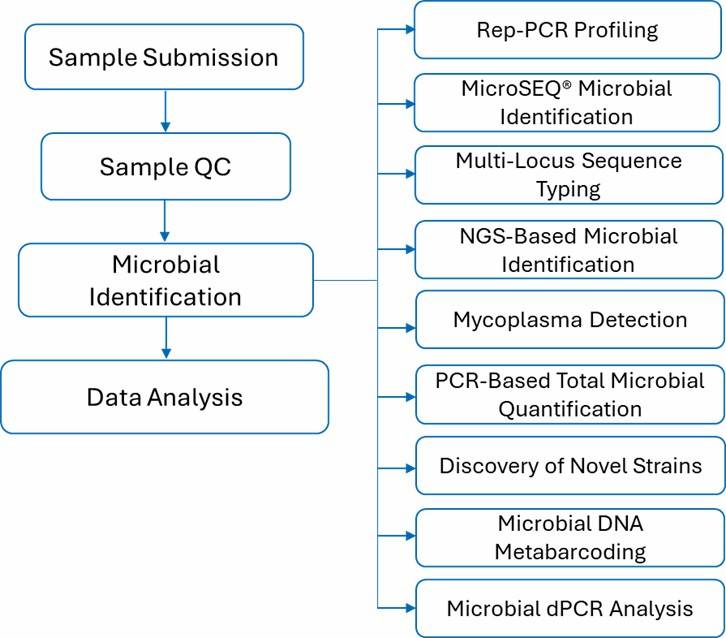
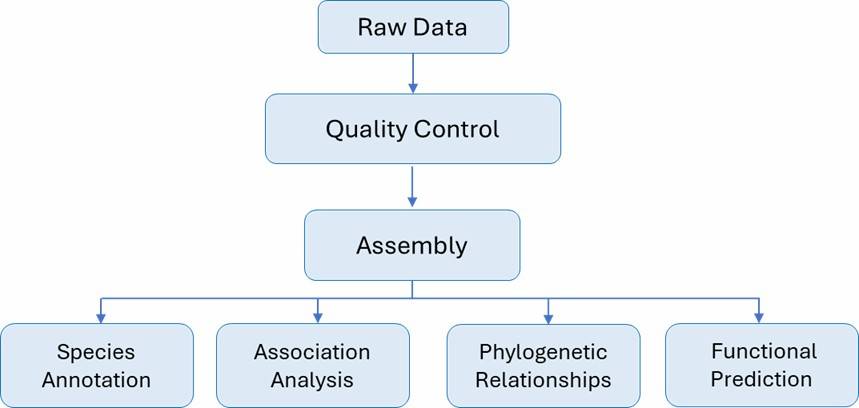

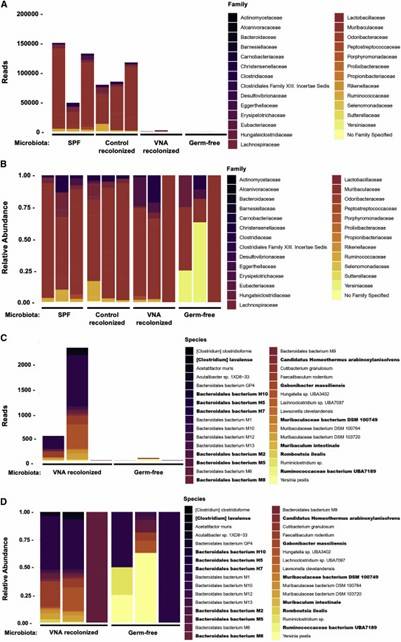 Fig 1. Bacterial species obtained from metagenomic sequencing.
Fig 1. Bacterial species obtained from metagenomic sequencing.