Importance of Functional Microbial Ecology
Microorganisms play an important role in biogeochemical cycles and soil nutrient reposition, They can also be a valuable indicator of soil quality, and directly affect crop development and crop health. Over the past decades, scientists have been trying to clarify questions about the diversity of species in relation to ecosystem functions. A range of biochemical, physiological, molecular, microscopic, and isotopic techniques are now available to identify and study various functional groups of microflora and their functions. The progress has now reached to a level that many technologies could be used for a reproducible, reliable assessment of the taxonomic and functional diversity and function of microbiota. Functional microbial ecology is a field to comprehensively determine population dynamics through functional diversity to the molecular regulation of key processes, which has been greatly powered by the next-generation sequencing (NGS) and bioinformatics analysis.
Accelerate Research and Practice in Functional Microbial Ecology
We are dedicated to exploring the diversity and evolution of functional microbial groups in diverse environments utilizing NGS and long-read sequencing technologies. According to your needs, we can design optimal, validated universe primers targeting functional genes or housekeeping genes (such as 16S rRNA, nirK, nirS, nosZ, LSU, amoA), and construct custom libraries for high-throughput sequencing. We can also apply this strategy to more refined classification and evolutionary research on functional microbial groups.
The Main Research Directions
- Molecular approaches to microbial ecology.
- Evaluate the microbial structure, evolution, and distribution in various environments.
- New strategies for management of plant pathogenic soil microorganisms.
What Can We Do?
- 1. Designed and validated universal primers for amplification of certain functional genes or housekeeping genes
- 2. Reveal the diversity of certain functional microbial groups in the sample
- 3. Evolutionary studies of certain groups of microbes
Note: Our service is for research use only, and not for therapeutic or diagnostic use.
Detectable Objects
Environmental samples, especially soil samples.
Detectable Microorganisms
| Functional microorganisms | The gene to be sequenced | Primers | Sequencing platform |
|---|---|---|---|
| Methanogens | 16S rRNA gene | Arch519R / Arch915R | MiSeq 2×300 |
| Sulfate-reducing bacteria (SRB) | dsrB | dsrF / dsrR | MiSeq 2×300 |
| Ammonia-oxidizing bacteria (AOB) | 16S rRNA gene | CTO189f / CTO653r | MiSeq 2×300 |
| Ammonia oxidizing bacteria (AOB) | amoA | amoA-1F / amoA-2R | MiSeq 2×300 |
| Ammonia-oxidizing archaea (AOA) | amoA | Arch amoA F / Arch amoA R | 454 |
| Nitrite oxidizing bacteria (NOB) | 16S rRNA gene | FGPS1269 / FGPS872 | MiSeq 2×300 |
| nxrA | nxrA F / nxrA R | MiSeq 2×300 | |
| Anammox (AMX) | 16S rRNA gene | Amx368 / Amx820R | MiSeq 2×300 |
| Denitrifying bacteria | narG | narG1960f / narG2650r | 454 |
| nirK | F1aCu / R3Cu | MiSeq 2×300 | |
| nirS | cd3aF / R3cd | MiSeq 2×300 | |
| nosZ | nosZF / nosZR | MiSeq 2×300 | |
| Pavlova gyrans | LSU | F / R | MiSeq 2×300 |
| Nitrogen-fixing bacteria | cbbL | 595F / 1387R | 454 |
| cbbM | F / R | MiSeq 2×300 | |
| Nitrogen-fixing bacteria | nifH | PoIF / PoIR | MiSeq 2×300 |
Detection Methods
The next-generation sequencing, third-generation sequencing, gene chip analysis, SNP genotyping, real-time qPCR, Denaturant Gel Gradient Electrophoresis (DGGE), etc.
Technical Platforms
We are equipped with Illumina Hiseq/Miseq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time qPCR, clone libraries, and other detection platforms.
Sample Requirements
-

- DNA amount > 300 ng, concentration > 10 ng/ul, and OD260/280 = 1.8-2.0.
- Please ensure DNA is non-degraded, and do not freeze or thaw the sample repeatedly during sample preservation.
- Please use enough ice bags or dry ice for sample submission.
Workflow
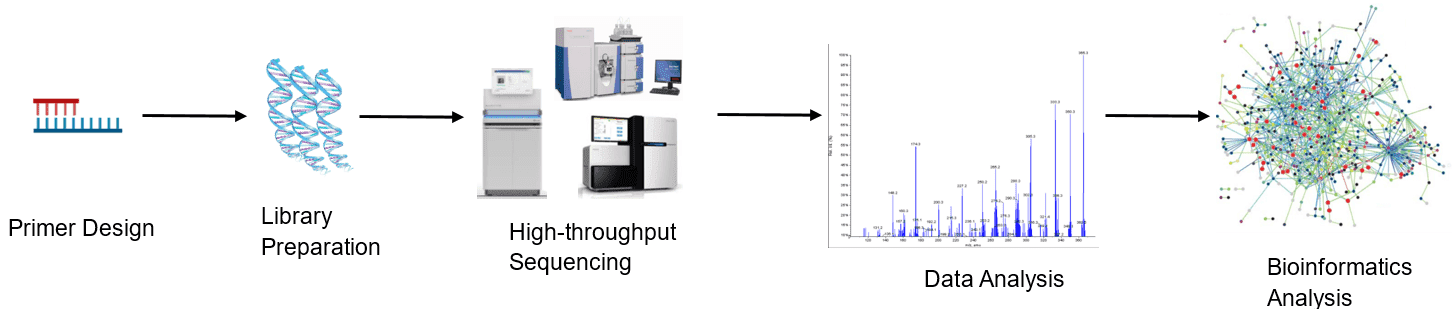 Figure 1. High-throughput sequencing analysis process.
Figure 1. High-throughput sequencing analysis process.

Bioinformatics Analysis
| Basic Analysis | Routine Analysis (According to Customer Requirements) | Advanced Data Analysis |
|---|---|---|
| Sequence Filtering and Trimming | Heatmap | Phylogenetic Tree |
| Sequence Length Distribution | VENN | LDA-Effect Size (LEfSe) |
| OTU Clustering and Species Annotation | Principal Components Analysis (PCA) | Network Analysis |
| Diversity Index | Microbial Community Structure Analysis | Correlation Analysis |
| Shannon-Wiener Curve | α Diversity Index Analysis | |
| Rank-Abundance Curve | Matastats Analysis | |
| Rarefaction Curve | Weighted Unifrac test | |
| Multiple Contrast | CCA/RDA Analysis | |
| Heatmap | ||
| Principal Components Analysis (PCA) |

