Backgrounds
Avian infectious laryngotracheitis virus (ILTV), or Gallid herpesvirus 1, is a herpesvirus causing severe respiratory disease in chickens with high economic impact. First found in the USA and later in China, it can cause significant mortality. The virus has a 150 kb DNA genome encoding about 80 proteins. Despite some identified virulence factors, key details remain unknown. This study compares the genomes and pathogenicity of three recent ILTV strains from China.
Methods
Sample preparation:
-
LMH cells
- ILTV
- DNA extraction
Data Analysis:
-
Sequence assembly
- Synteny analysis
- Sequence alignment
- Phylogenetic analysis
- Statistical analysis
Results
The complete genomes of three ILTV strains—SH2016, SH2017, and GD2018—were sequenced and deposited in GenBank. The genomes are nearly identical, with SH2017 and GD2018 sharing 99.9% similarity, while SH2016 is 99.7% similar to the others. All strains have 82 ORFs, with 31 proteins being identical. Notably, SH2016 differs from SH2017 and GD2018 in eight specific proteins.
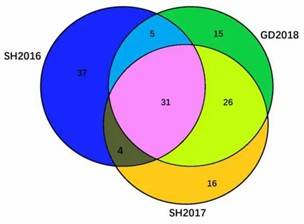 Figure 1. Diagram of proteins of three ILTVs.
Figure 1. Diagram of proteins of three ILTVs.
Phylogenetic analysis of 20 ILTV strains reveals that SH2016, SH2017, and GD2018 fall into distinct sub-lineages. SH2016 clusters with Russian and Australian vaccine strains, indicating possible evolution from these vaccines. SH2017 and GD2018, showing 99.9% similarity, are closely related to U.S. vaccine strains, suggesting a different evolutionary path.
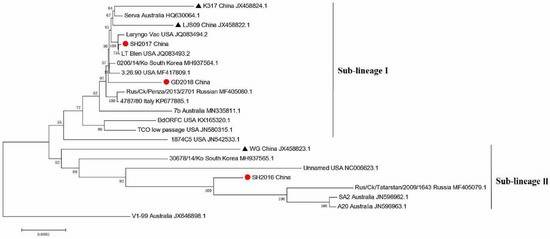 Figure 2. Phylogenetic tree of ILTVs
Figure 2. Phylogenetic tree of ILTVs
Conclusions
Three ILTV isolates, despite high genomic similarity, exhibit notable differences in replication and pathogenicity. SH2016 is more virulent, while SH2017 and GD2018 cause milder symptoms. Variations in specific viral proteins likely account for these differences.

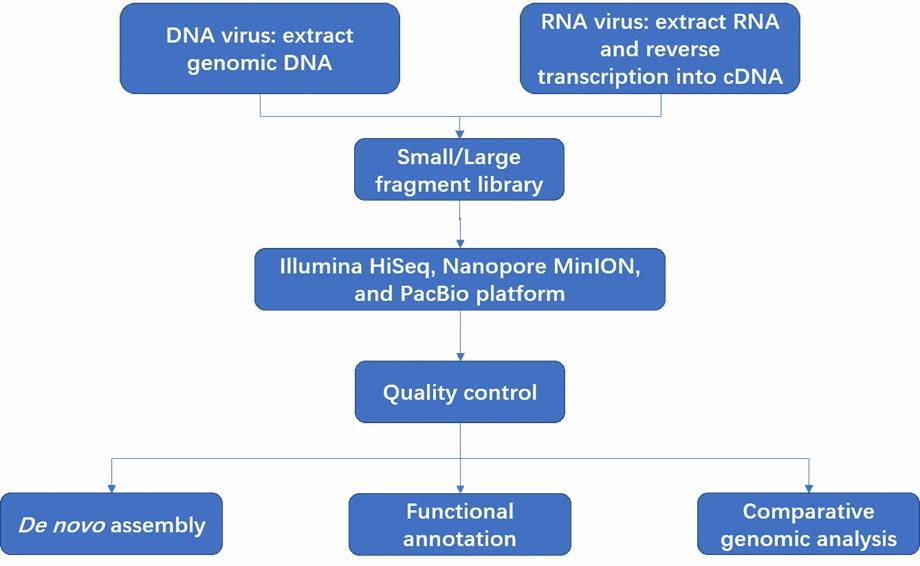
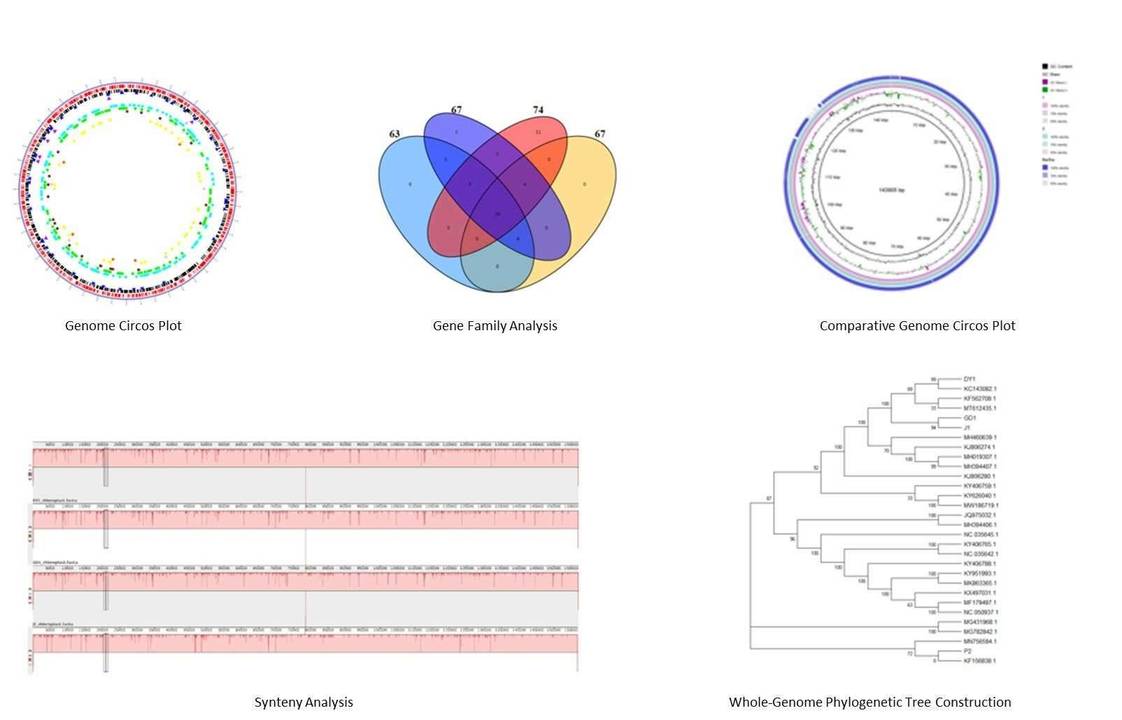

 Figure 1. Diagram of proteins of three ILTVs.
Figure 1. Diagram of proteins of three ILTVs. Figure 2. Phylogenetic tree of ILTVs
Figure 2. Phylogenetic tree of ILTVs