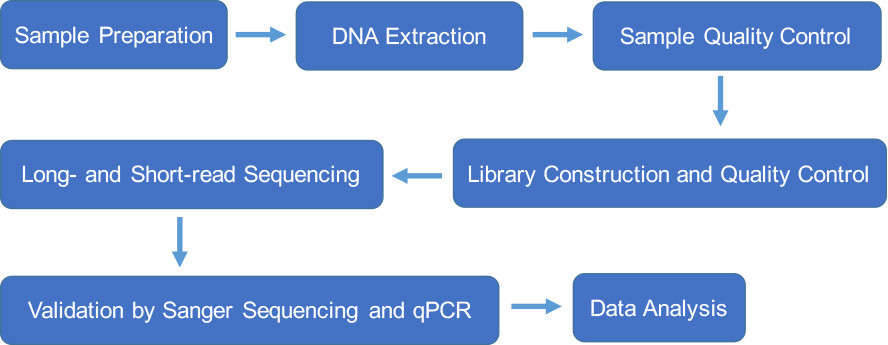
We utilize both long-read and short-read sequencing technologies for fungal whole genome de novo sequencing. Both PacBio and Nanopore platforms are superior long-read sequencing platforms that generate reads with tens of kilobases in length for better genome reconstruction. A single molecule of DNA can be sequenced without the need for PCR amplification or chemical labeling of the sample. De novo assembly from long reads can span the low complexity and repetitive regions and create accurate assemblies. Additionally, we perform fungal whole genome de novo sequencing on Illumina HiSeq, which is a short-read sequencing platform with high accuracy.
Fungal whole genome de novo sequencing, which is used to explore novel fungal populations, enables the fungal genomes reconstruction, functional analysis, and evolutionary studies. This technique can provide insight into the biological diversity of fungi and yeast, thereby expanding research into agricultural science, ecology, bioremediation, medical science, cosmetic science, bioenergy, and biotechnology industry. We also provide fungal whole genome resequencing to detect variants, explore gene functions, and reconstruct phylogenetic trees.