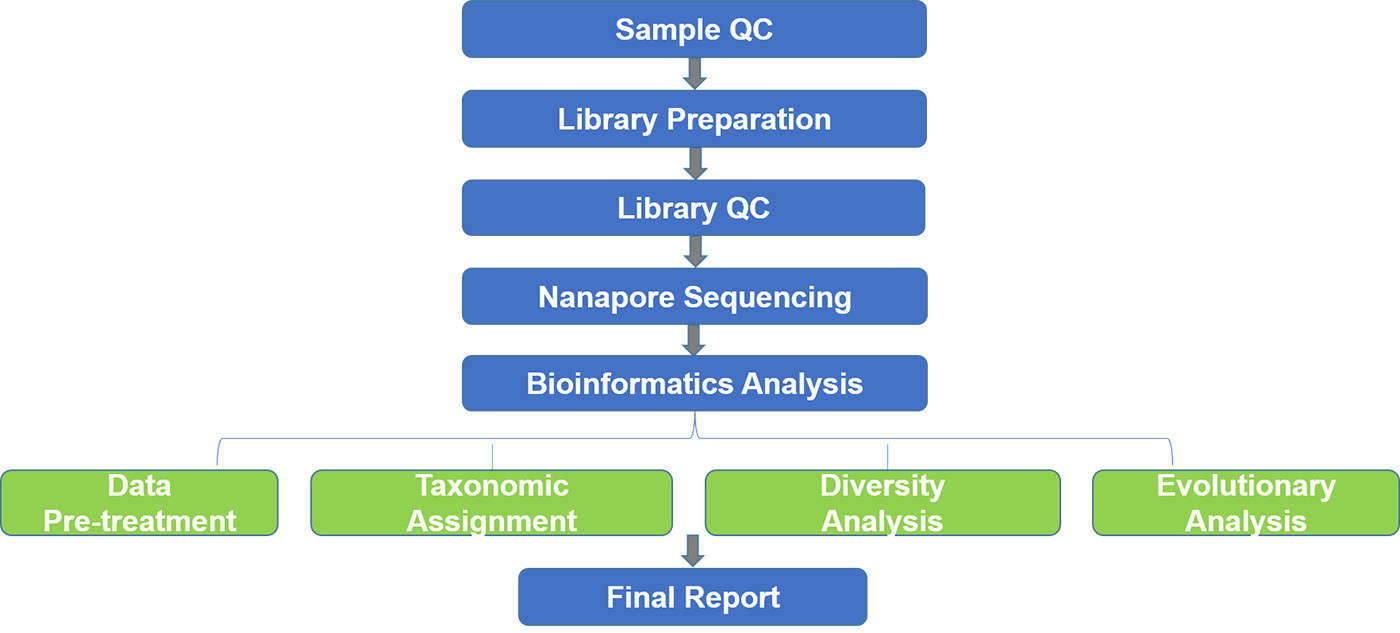
The 16S rRNA gene is present in all archaea and bacteria, and consists of conserved and highly variable regions, making it ideal for the identification of prokaryotes. The 18S rRNA gene and internally transcribed spacer (ITS) are present in all eukaryotes and are suitable for eukaryotic identification and diversity analysis. Nanopore sequencing, a long-read sequencing technology, has obvious advantages in generating a more accurate profile of the microbiome than short-read sequencing technologies. It holds the promise of a microbiology revolution through generating real-time, long reads, with continuously increasing accuracy. Our nanopore-based 16S/18S/ITS sequencing provides rapid and accurate prokaryotic identification analysis with a relatively simple workflow and no need for DNA purification.
Nanopore-based 16S/18S/ITS sequencing is a novel molecular approach, with a more comprehensive and rapid process compared to the traditional culture-based bacterial identification, for the taxonomic characterization of microbial communities. In microbiology, nanopore-based full-length 16S/18S/ITS sequencing can be used to diagnose bacterial infections and monitor outbreaks as acute infectious diseases remain one of the major causes of human diseases with high mortality. It can also be used to monitor and improve human health through gut or skin microbial diversity analysis. Nanopore-based 16S/18S/ITS sequencing can be applied to understand the role of microbes in agricultural, environmental, and health-related settings.