Many microorganisms, including bacteria, viruses and fungi, coexist in the lungs of humans or animals and constitute the respiratory microbiota. It is generally accepted in the scientific community that the lung or respiratory microbiota is less biologically abundant, scarcer, and associated with mucosal surfaces than the relatively stable intestinal community. In this dystrophic microbial environment, high-throughput sequencing technologies can not only capture signals with exceptionally low DNA/RNA densities and characterize a variety of novel in samples but can also simultaneously study pathogen and host responses through molecular approaches such as microbial metagenomics and transcriptomics.
CD Genomics is dedicated to breaking down the limitations of lung microbiome research by NGS and long-read sequencing platforms, processing or speed to help you generate and comprehend high-dimensional data.
 Request a Quote
Request a Quote
What can we do?
1. Characterization of the respiratory microbiome
2. Quantitation of the respiratory microbiome
3. Revealing a wealth of information about community composition and dynamics
Note: Our services are for research use only, not for disease diagnosis and treatment.
Detectable Objects
Sputum, alveolar lavage fluid, bronchial brushings and lung tissue, and other lung microbiome sample types
Detectable Microorganisms
Bacteria, fungi, viruses, etc.
Technology Platform
We are equipped with Illumina Hiseq/Miseq, PacBio SMRT systems, Nanopore systems, PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time qPCR, and other detection platforms.




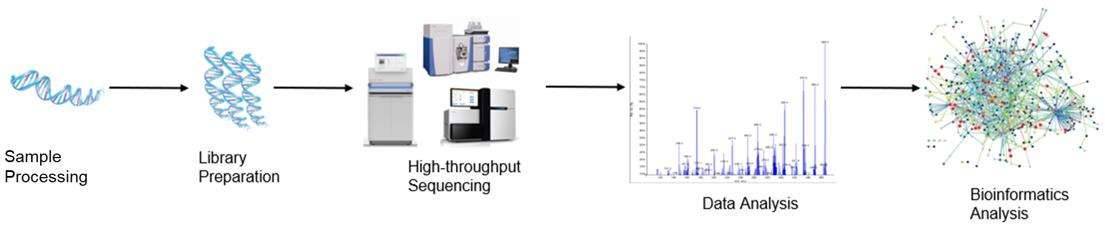 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process