What Is mNGS
Metagenomic Next-Generation Sequencing (mNGS) involves a comprehensive analysis of microbial and host genetic material (DNA and RNA) in a given sample. The application of microbial metagenomic sequencing (mNGS) technology enables the detection of a broader spectrum of pathogens. Presently, mNGS is swiftly transitioning from scientific research to clinical diagnostic applications, excelling in the identification of rare, emerging, challenging-to-detect, and mixed infections.
If traditional pathogen-specific detection methods are likened to "fishing" for specific targets, mNGS can be compared to "casting a wide net." Since 2008, extensive research from over 20 countries has demonstrated the broad utility of mNGS. Its applications span pathogen detection and identification, antibiotic resistance analysis, microbiome characterization, assessment of anti-infective treatment efficacy, human host gene expression transcriptomics, and oncology, among other areas.
Distinctive Features of mNGS Sequencing Technology
The predominant approach of traditional methods for identifying infectious disease-causing microorganisms involves making assumptions, conducting experiments, and subsequently validating the findings. The identified range of microbial species is limited with these methods. In contrast, mNGS, which does not rely on predefined assumptions, holds the potential to detect all types of microorganisms. A comparative analysis of different detection technologies reveals the following:
| Method | Advantages | Disadvantages |
|---|---|---|
| PCR | Simple, fast, cost-effective, quantitative detection, high accuracy. | Relies on subjective assumptions, detects known microorganisms, dependent on primer amplification (which may not always be reliable), limited to a small portion of the microbial genome. |
| Cultivation Method | Adaptable to larger sample volumes, cost-effective, considered the "gold standard" for comparative studies. | Sensitivity limited by antibiotic use during cultivation, difficulty in culturing some microorganisms, inability to detect viruses, long detection cycles. |
| Mass Spectrometry: | Simple process, high specificity and accuracy, capable of high-throughput analysis. | Relies on cultured positive colonies, limited to about 1000 known microorganisms, some pathogens cannot be identified to the species level, qualitative rather than quantitative. |
| Immunological Detection | Timely, short cycles, cost-effective, simple operation, detects chlamydia, mycoplasma, viruses, etc. | Poor sensitivity and specificity, not all pathogenic microorganisms have corresponding antibodies, susceptible to false positives, potential for window periods. |
| mNGS | Objective detection, independent of assumptions, detects any microorganism, discovers new pathogens, quantitative nucleic acid detection, can examine any part of the genome. | Results influenced by host background microbiome, high sample requirements, susceptible to contamination from other sites and environmental microorganisms, less timely than PCR nucleic acid detection, higher cost. |
| tNGS | Unaffected by the human genome and background bacteria, enhanced performance in detecting antibiotic resistance genes, higher cost-effectiveness, sensitive detection of low-abundance pathogens. | Unable to detect unknown microorganisms. |
Pathogenic mNGS Service
CD Genomics Launches Comprehensive Pathogenic Sequencing Solution, expanding the scope of detection, optimizing cost-effectiveness, enhancing efficiency, and maximizing productivity.
mNGS Workflow
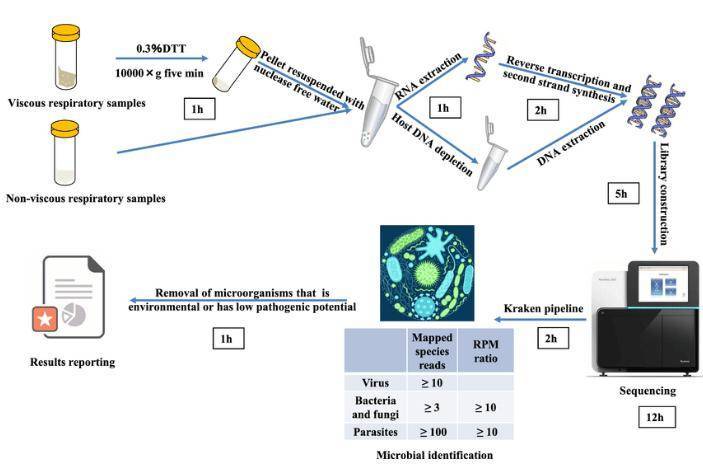 Complete mNGS assay workflow. (Yukun He et al,. 2022)
Complete mNGS assay workflow. (Yukun He et al,. 2022)
Utilizing metagenomic next-generation sequencing (mNGS) technology for the detection of pathogenic microorganisms typically involves six sequential steps: sample collection, nucleic acid extraction, construction of standardized sequencing libraries from the extracted nucleic acids, high-throughput sequencing, bioinformatics analysis to identify the pathogenic bacteria, and interpretation of the findings in the report.
Sample Requirment
| Sample Type | Sample Volume | Sample Type | Sample Volume |
|---|---|---|---|
| Peripheral Blood | ≥4ml | Cerebrospinal Fluid, Other Body Fluids | ≥1ml |
| Bone Marrow Fluid | ≥1ml | Urine | ≥1ml |
| Nasopharyngeal Swab, Wound Exudate | ≥1 swab | Aqueous Humor | ≥0.1ml |
| Bronchoalveolar Lavage, Abscess Fluid | ≥5ml | Vitreous Humor | ≥0.5ml |
| Sputum, Pleural/Peritoneal Fluid, Joint Effusion | ≥1ml | Bile | ≥2ml |
| Tissue | ≥ 3*3*3 mm |
Product Advantages:
- No need for ex vivo cultivation and purification.
- Detects drug-resistant genes.
- Simultaneously detects DNA and RNA.
- Wide coverage, capable of detecting over 15,000 pathogenic microorganisms, including bacteria, fungi, viruses, parasites, mycoplasma/chlamydia, rickettsia, and spirochetes.
- Efficient and accurate comprehensive analysis reports.
In addition, CD Genomics leverages NGS technology to provide targeted NGS (tNGS) and comprehensive pathogen whole genomics sequencing services to global pathogen research projects.

