Explore Complex Microbiomes with 5R 16S Microbial Diversity Sequencing
Service Overview
Achieve species-level microbial profiling even from degraded or low-biomass samples
The 5R 16S Microbial Diversity Sequencing service offers comprehensive and high-resolution microbial community profiling by targeting five hypervariable regions (V2, V3, V5, V6, V8) of the 16S rRNA gene. Through multiplex PCR amplification combined with the advanced Short MUltiple Regions Framework (SMURF) algorithm, this method provides enhanced taxonomic resolution and coverage, especially for challenging sample types such as clinical tissues, FFPE blocks, and blood.
For standard environmental or fecal microbiome studies, our Bacterial 16S rRNA Sequencing service based on the V3-V4 region remains a widely used baseline solution.
Technical Principle
Unlike conventional single- or dual-region 16S sequencing, 5R 16S simultaneously amplifies five distinct variable regions of the 16S rRNA gene, covering approximately 68% of the full-length sequence. The resulting data is processed using the SMURF algorithm, enabling reconstruction of microbial profiles with species-level resolution and broad taxonomic coverage.
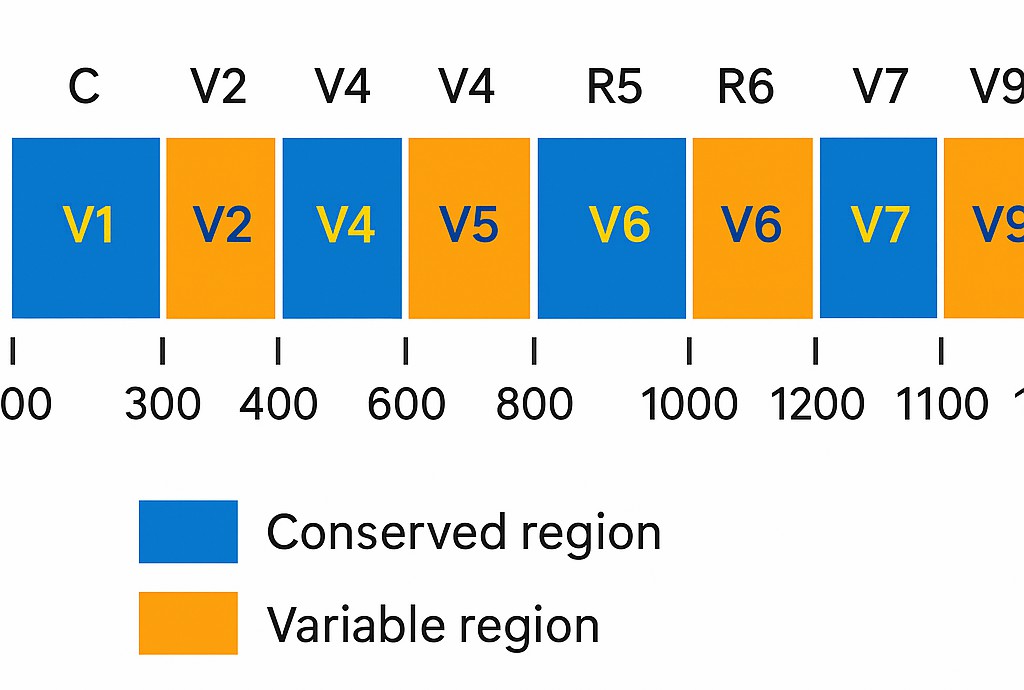 Figure 1. Graphic representation of bacterial 16S rRNA gene, showing conserved and variable regions
Figure 1. Graphic representation of bacterial 16S rRNA gene, showing conserved and variable regions
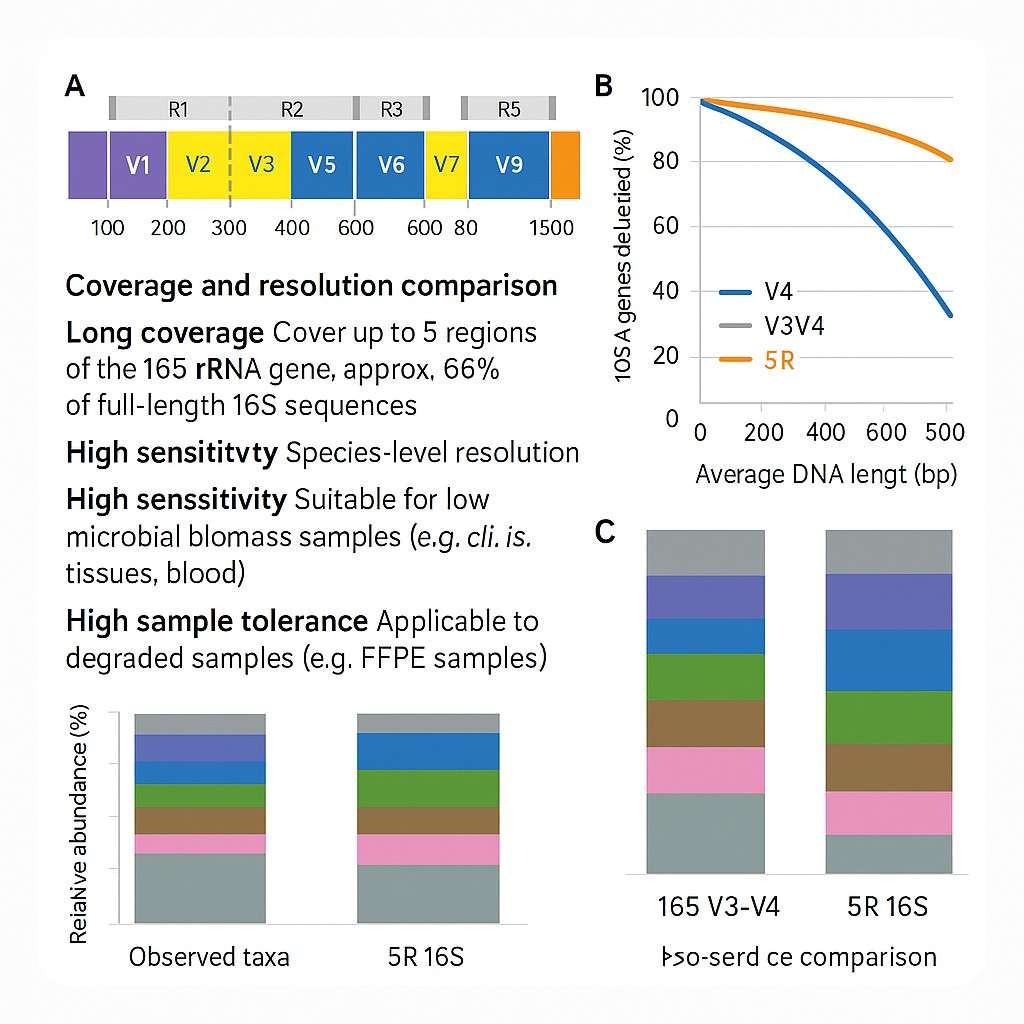 Figure 2. Comparison of taxonomic resolution and coverage between single-region, dual-region, and 5R 16S sequencing approaches
Figure 2. Comparison of taxonomic resolution and coverage between single-region, dual-region, and 5R 16S sequencing approaches
For projects requiring maximum read length or rare taxon recovery, consider our Full-Length 16S/18S/ITS Sequencing service using long-read technologies.
Key Features and Applications
| Feature |
Description |
| Extended Coverage |
Targets five hypervariable regions (V2, V3, V5, V6, V8), covering ~68% of 16S gene length. |
| Species-Level Resolution |
Enables precise taxonomic classification down to the species level. |
| High Sensitivity |
Ideal for low-biomass samples such as tumor tissue, blood, or lung samples. |
| Robust to Degraded DNA |
Compatible with partially degraded samples, including FFPE-preserved tissues. |
For metagenomic profiling of ultra-low biomass environments such as sterile tissues, cerebrospinal fluid, or even cell-free DNA, we also recommend 2bRAD-M Analysis for its robustness and species-level output.
Technical Comparison: 5R 16S vs. V3-V4 Sequencing
| Parameter |
16S V3-V4 |
5R 16S |
| Target Samples |
High-biomass (e.g., feces) |
Low-biomass (e.g., tumors, blood) |
| Compatibility with FFPE |
Low |
High |
| Sequencing Platform |
Next-Generation Sequencing |
Next-Generation Sequencing |
| Variable Regions Sequenced |
2 (V3-V4) |
5 (V2, V3, V5, V6, V8) |
| Bioinformatics Pipeline |
QIIME2 + DADA2 |
SMURF |
| Taxonomic Resolution |
Genus level |
Species level |
| Sensitivity |
Standard |
Higher |
Workflow
Advanced Bioinformatics Analysis Summary
| Bioinformatics Analysis |
Problem Solved |
| Species annotation |
OTU clustering, taxonomic classification |
| Diversity analysis - α diversity |
Evaluates species richness within a single microbial ecosystem |
| Diversity analysis - β diversity |
Assesses differences in microbial communities between environments |
| Meta-analysis |
Compares feature abundance across different datasets |
| Evolutionary analysis |
Builds phylogenetic tree to determine evolutionary relationships |
| Functional analysis - Intestinal Microflora |
Identifies gut bacteria, predicts disease associations, aids in treatment strategies |
| Functional analysis - PICRUSt |
Predicts functional capabilities of microbial communities |
| Functional analysis - FAPROTAX |
Links microbial taxa to biogeochemical cycling processes |
| Functional analysis - Tax4fun |
Infers microbial functional profiles based on KEGG pathways |
| Functional analysis - FunGuild |
Classifies ecological roles of fungal and eukaryotic OTUs |
| Correlation analysis - CCA |
Reveals microbial-environment factor correlations using ordination |
| Correlation analysis - VPA |
Partitions variance among factors influencing community structure |
| Correlation analysis - Network analysis |
Analyzes microbial interaction patterns and co-occurrence networks |
Sample Requirements
Please ship all samples on dry ice to ensure integrity.
| Sample Type |
Requirements |
| FFPE Samples |
≥20 μm thick slices; ≥6 slices per sample |
| Animal Tissue |
Fresh muscle (0.1-0.3 g); snap-freeze in liquid nitrogen and store at -80 °C |
| Blood Samples |
0.5-1 mL in EDTA tubes; gently invert to mix; transfer to sterile 2 mL microcentrifuge tubes; store at -80 °C (Do not ship glass collection tubes) |
| DNA Samples |
≥15 μL volume; concentration >20 ng/μL |
Case Dispaly
Case Study: Mouse Lung Tissue Microbiome
To evaluate the performance of 5R 16S sequencing, we analyzed the same mouse lung tissue sample using both 16S V3-V4 dual-region sequencing and 5R 16S. The top six microbial taxa identified by the V3-V4 method included several potential environmental contaminants. In contrast, 5R 16S sequencing provided a more accurate and comprehensive profile of the actual microbial population present in the lung tissue.
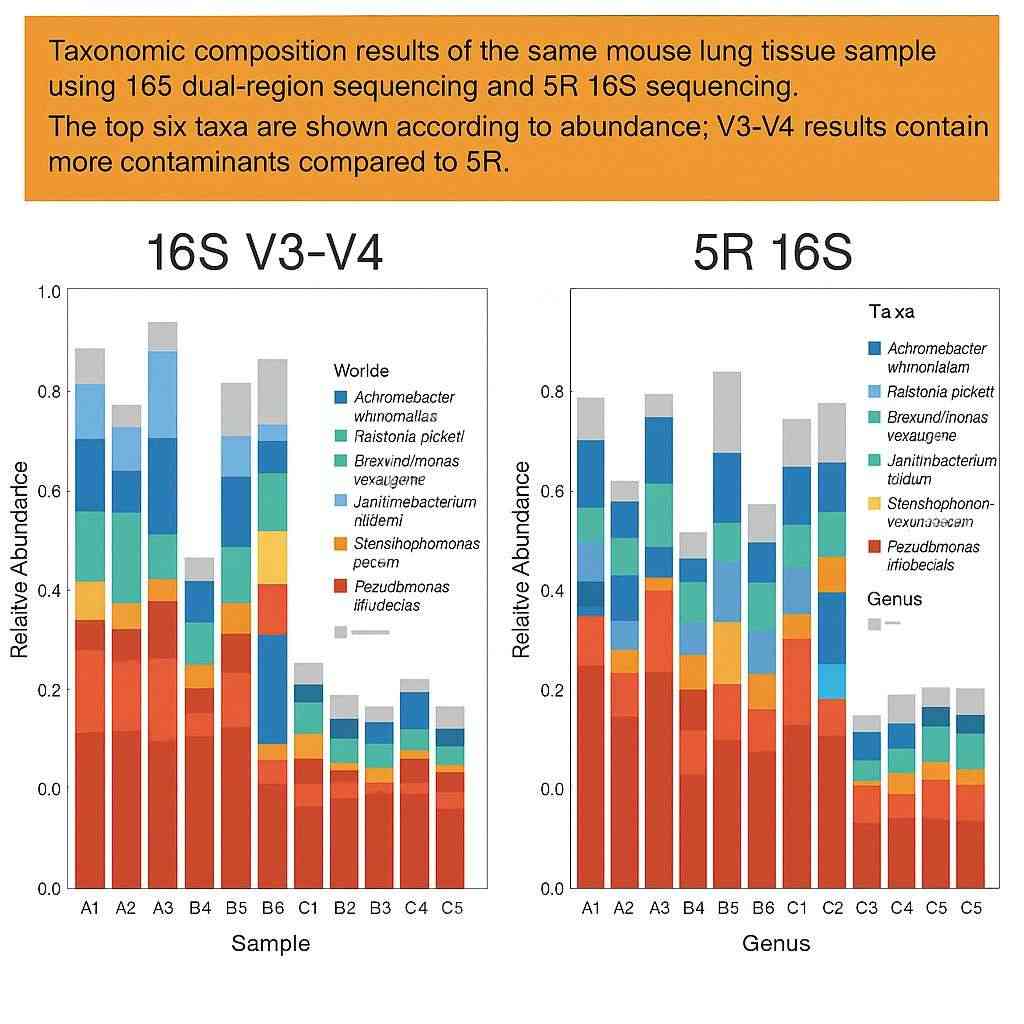 Figure 3. Comparison of microbial composition results:
Figure 3. Comparison of microbial composition results:
Left: 16S V3-V4 sequencing | Right: 5R 16S sequencing
To complement sequencing data with spatial localization, we offer custom 16S and 18S FISH probe design, synthesis, and in situ detection. This is especially useful in histological studies where localization within tissues is critical.
Explore our FISH Probe Design & Detection Service to visualize microbial distributions directly in tissue sections.
Why Choose Us?
Creative Proteomics combines robust laboratory protocols with advanced informatics to provide accurate, sensitive, and reproducible 16S microbiome sequencing results for even the most difficult sample types. With our 5R 16S Microbial Diversity Sequencing service, you'll uncover microbial insights that conventional methods might miss.
FAQ
Frequently Asked Questions (FAQs)
- 1. What advantages does 5R 16S sequencing offer over conventional V3-V4 sequencing?
- 2. Is this method suitable for FFPE samples or old tissue archives?
- 3. How much DNA is required for library construction?
- 4. Can I use this method for blood, tumor tissue, or cerebrospinal fluid?
- 5. Do you provide downstream analysis and visual reports?
- 6. How is this different from full-length 16S sequencing?
- 7. Can I combine this with other services like ITS or 2bRAD?
- 8. Do you help with publication or result interpretation?
References
- Nejman D, et al. Science. 2020;368(6494):973-980. https://doi.org/10.1126/science.aay9189
- Fuks G, et al. Microbiome. 2018;6(1):17. https://doi.org/10.1186/s40168-017-0396-x
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Figure 1. Graphic representation of bacterial 16S rRNA gene, showing conserved and variable regions
Figure 1. Graphic representation of bacterial 16S rRNA gene, showing conserved and variable regions  Figure 2. Comparison of taxonomic resolution and coverage between single-region, dual-region, and 5R 16S sequencing approaches
Figure 2. Comparison of taxonomic resolution and coverage between single-region, dual-region, and 5R 16S sequencing approaches
 Figure 3. Comparison of microbial composition results:
Figure 3. Comparison of microbial composition results: