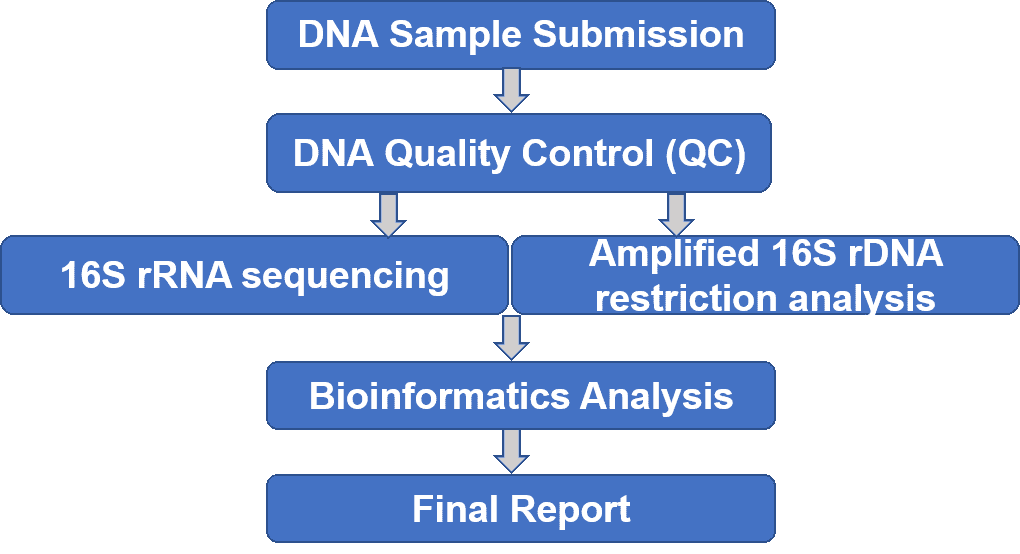
We provide comprehensive diversity analysis of actinomycetes using molecular methods including 16S ribosomal RNA (rRNA) gene sequencing and amplified 16S ribosomal DNA restriction analysis (ARDRA). 16S rRNA sequencing generally involves the selective PCR amplification using appropriate primers that target the16S rRNA gene, followed by next/third generation sequencing. ARDRA is a reliable identification method based on restriction endonuclease (such as AluI, HaeIII, MspI, etc.) digestion of the amplified bacterial 16S rDNA. Our service can help with strain assignment and new species discovery.
Since actinomycetes have been considered as the richest source of secondary metabolites such as antibiotics, diversity analysis of actinomycetes aims to investigate the diversity of antibiotic-producing actinomycetes and accelerate the development of a novel antibiotic drugs. Diversity analysis of actinomycetes can also confirm the diagnosis of actinomycosis. The importance of this analysis has been demonstrated in areas such as drug discovery and development, agricultural production, petroleum exploration, and industrial manufacturing, etc.