Introduction to Biofilm Microorganisms
Biofilms refer to the bacterial communities that attach to surfaces. They contribute to the wellbeing and resilience of ecosystems via their important roles in biogeochemical cycling, their interactions with each other and their provision of chemicals. Although aquatic biofilms are important to the health of marine ecosystems, they also cause deleterious outcomes like biocorrosion, biofouling and antibiotic resistance. When it comes to human health, microbes can form biofilms on any surface in the human body and cause resistance to antimicrobial agents, triggering persistent and chronic bacterial infections. Biofilm infections have sharply come into prominence in the last few decades. Next-generation sequencing (NGS) is a powerful tool when studying the biofilm microbiomes. Genomic studies have revealed the complexity and diversity of the biofilms.
Accelerate Research and Practice in Microbial Diversity in Biofilms
We provide advanced genomics methods to explore the microbial diversity and structure of biofilm samples, and discover previously undetected microbial species and predict their roles in biofilm formation, human health or environments. Our solutions include real-time PCR, 16S/18S/ITS sequencing, metagenomics, metatranscriptomics and powerful bioinformatics analysis. Based on Illumina Hiseq/Miseq, PacBio and Oxford nanopore sequencing platforms, we are dedicated to ensuring an accurate & comprehensive microbial profiling and elaborate bioinformatics analysis pipeline with standardized laboratory methods.
The Main Directions
- Exploration of microbial diversity and structure of biofilms
- Functional core analysis and discovery of persistent and distinct functional cores
- Studies of community succession and microbe-host interactions
- Diagnosis and treatment of different biofilm infections
What Can We Do?
- 1. Identification of biofilm microorganisms and taxonomic profiling
- 2. Quantitation of biofilm microorganisms
- 3. Functional core analysis and exploration of novel functional potential
Note: our service is for research use only, not for diagnosis.
Detectable Objects
Artificial biofilms, marine microbial biofilms and other biofilms on any surface.
Detectable Microorganisms
Archaea, bacteria, eukaryotes, etc.
Technical Platforms
We are equipped with Illumina Hiseq/Miseq, PacBio Sequel, Oxford MinIONTM, Oxford GridIONTM, PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time qPCR, clone library and other platforms.
Sample Requirement
-

- DNA sample: DNA ≥ 300 ng, concentration ≥ 10 ng/ul, OD260/280 = 1.8 - 2.0
- Make sure the DNA is intact and non-degraded, avoiding repeated freezing and thawing cycles.
- Please prepare enough dry ice or ice packs to ensure low temperature during sample delivery.
Workflow
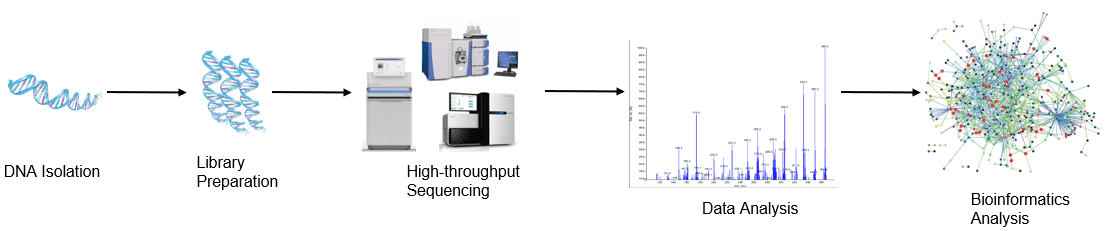 Figure 1. High throughput sequencing analysis process
Figure 1. High throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
Our bioinformatics analysis pipeline is flexible to your needs.
| Basic Analysis | Routine Analysis (According to Customer Requirements) | Advanced Data Analysis |
|---|---|---|
| Read QC | Heatmap | Phylogenetic Tree |
| Sequence Length Distribution | VENN | Functional Core Analysis |
| OTU Clustering and Taxonomic Profiling | Principal Components Analysis (PCA) | LDA-Effect Size (LEfSe) |
| Diversity Index | Microbial Community Structure Analysis | Network Analysis |
| Shannon-Wiener Curve | α Diversity Index Analysis | Correlation Analysis |
| Rank-Abundance Curve | Matastats Analysis | |
| Rarefraction Curve | Weighted Unifrac test | |
| Multiple Contrast | CCA/RDA | |
| Heatmap | ||
| Principal Components Analysis (PCA) |
References
- Zhang W, Ding W, Li Y X, et al. Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nature communications, 2019, 10(1): 517.
- Sauer K. The genomics and proteomics of biofilm formation. Genome biology, 2003, 4(6): 219.
- Bjarnsholt T. The role of bacterial biofilms in chronic infections. Apmis, 2013, 121: 1-58.