The Importance of Microbial Biomarker Discovery
The commensal microbes associated with humans or animals contribute to the host health as they play a vital role in shaping host immunity and preventing infectious disease. Given the widespread presence of antimicrobial resistance, promoting healthy microbiota in humans and animals became a global issue. Biomarker discovery has proven to be one of the most powerful and broadly applicable approach to translate molecular and genomic data into practice. However, the bacterial genome is highly dynamic even within strains of the same genus. Therefore, the identification of useful biomarkers in mixed microbial communities is very challenging. High-resolution techniques such as next-generation sequencing (NGS) and long-read sequencing are necessary for effective microbial biomarker discovery. Genomic methods can also be used to systematically discover potential biomarker genes through deep sequencing, literature-based data mining, and comparative genomics.
Our Solution for Microbial Biomarker Discovery
We are dedicated to providing the highest-quality microbial genomics services to help you investigate microbe-microbe and host-microbe interactions within the human/animal microbial communities. We utilize next-generation sequencing (NGS) and long-read sequencing technologies to characterize microbial communities. We provide (full-length) 16S/18S/ITS sequencing and metagenomics for rapid detection of bacteria, archaea, fungi, and viruses, and a more comprehensive combination of microbial genomics services for further characterization of microbes, genes, or pathways of interest.
Main Research Directions
- Microbiome-wide association studies linking dynamic microbial consortia with disease.
- Discovery, validation, and identification of microbial biomarkers.
What Can We Do?
- 1. Microbial diversity analysis using (full-length) 16S/18S/ITS sequencing and metagenomics
- 2. Discovery of microbial biomarkers using microbial diversity analysis and metatranscriptomics
- 3. Further characterization of bacteria and fungi using microbial whole-genome sequencing and microbial epigenomics
Note: Our service is for research use only, not for disease diagnosis or treatment.
Detectable Objects
Bacteria, fungi, protozoan, viruses.
Detection Methods
Next-generation sequencing (NGS), long-read sequencing, quantitative PCR (qPCR), and other molecular biology techniques.
Detection Platforms
Illumina HiSeq/MiSeq, Nanopore MinIONTM, PacBio RSII, PacBio Sequel, Applied Biosystems real-time PCR platforms, et al.
Sample Requirement
-

- DNA sample: 1.8 < OD260/280 < 2.0 (without degradation or contamination)
- RNA sample: RIN ≥ 7.0, 28s/18s ≥ 1.5 (without degradation or contamination)
Workflow
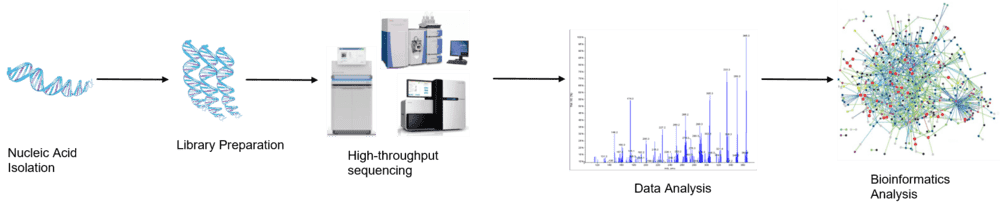 Figure 1. The high-throughput sequencing analysis process.
Figure 1. The high-throughput sequencing analysis process.
Bioinformatics Analysis
Our bioinformatics analysis pipeline is flexible to your needs.
| Taxonomic Assignment | Species annotation |
| Rarefaction Curve | |
| Shann-Wiener Curve | |
| Rank Abundance Curve | |
| Statistical Analysis | Heatmap |
| VENN | |
| Principal Components Analysis (PCA) | |
| Comparative Analysis | RDA/CCA |
| PCA/PCoA | |
| LEfSe | |
| Network Analysis | |
| Functional analysis | BLASTX, KEGG, eggNOG, CAZy, CARD, ARDB etc. |
| PICRUSt | |
| TAX4Fun | |
| Phylogenetic Analysis | (Un)Weighted Unifrac |
| Phylogenetic Trees |
References
- Yan X, Gurtler J, Fratamico P, et al. Comprehensive approaches to molecular biomarker discovery for detection and identification of Cronobacter spp.(Enterobacter sakazakii) and Salmonella spp. Appl. Environ. Microbiol, 2011, 77(5): 1833-1843.
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome biology, 2011, 12(6): R60.